Locus name: Al0029
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 60Main product (bp): 1200
Comment on PCR:
F-primer: TGGGGAAAGAAAGACTACAAT
R-primer: AAAACCTAACCCGCAAAT
EST info
EST Accession number 1: C95227EST Accession number 2: DC892873
sequence-tagged site (STS seq)
STS:| AACAAGAAACAAAAACAAAAGAATGAAAAACAATTATTTTTGTTTTTGTAAAACAGAACA CGCTCTCCCCAAAAACCAACCACACCCACCCACTCACACACGCACACTCATGACTCGCTC GCCAAAAGAAACAAAAGCCTAATCAATCCGCAAACCCAATCACCACAAAACGACAATCTA AAAGCAGCACGCGTGCCCCCACCACCTTTACTACTTTCGCAGATTCCCGCCCTCTCTCTC TCTCTCTCTATCACCTTTATATGCAACAATGACTTTTTATTTTAATTTATTTTTATTTCT CTTTTTTGTTTTTTTCAATGAAAAGGGAGGAGGAGGTGGTGATTGTATCCAAGCAAACGG GCGGAACCCTAACGGAGCTCAGACGACGTCAGCTAACGACAGCTTAACTGTGACGGAACA CCAAGTGGCTAAGCTGATGGAAGAAGATATGGGGTCAGCCATGCAGTACTTACAAGGCAA AGGGCTTTGTCTAATGCCCATCTCACTTGCAACTGCTATATCGACTGCCACGTGTCACTC TAGGAACCCCATCATCAGCACCAGCAACAACAACAATAACAACGGCAATCCCCACCACAA TCCTTTGCTTCAGTCCAATGGTGAGGGCCCGACCTCACCTAGCATGTCAGTGCTGACTGT CCAGTCAGCAACTATGGGTAACGGTGGGGCTGATGGCTCCGTCAAAGATGCTGCTNCGTT TCAAGCCTTGATCACGGCGTNGNCGANTTATGANANGCTCGNCAAGTCTACNAAATAGGC TCCTAGAGNNGGACTCCCACGCGATTGGGAACTTGGCGATTACACACAAAGCAGAGAGCA NCAACAACTTTTGTTTTTTT |
STS Forward / Reverse: Foward read
STS:
| AAACAAAGATGAAACCATTTTTTGTTTNTAAACAGACACGTTTTCCCAAAAAACAACACA CCANCCANTCACACACGCACATTCATGATTGGTCGCCAAAGAAACAAAGCTTATCAATCC GCAAACCCAATCACCACAAAACGACAATCTANAAGCAGCACGCGTGCCCCCACCACCTTT ACTACTTTCGCAGATTCCCGCCCTCTCTCTCTCTCTCTCTATCACCTTTATATGCAACAA TGACTTTTTATTTTAATTTATTTTTATTTCTCTTTTTTGTTTTTTTCAATGAAAAGGGAG GAGGAGGTGGTGATTGTATCCAAGCAAACGGGCGGAACCCTAACGGAGCTCAGACGACGT CAGCTAACGACAGCTTAACTGTGACGGAACACCAAGTGGCTAAGCTGATGGAAGAAGATA TGGGGTCAGCCATGCAGTACTTACAAGGCAAAGGGCTTTGTCTAATGCCCATCTCACTTG CAACTGCTATATCGACTGCCACGTGTCACTCTAGGAACCCCATCATCAGCACCAGCAACA ACAACAATAACAACGGCAATCCCCACCACAATCCTTTGCTTCAGTCCAATGGTGAGGGCC CGACCTCACCTAGCATGTCAGTGCTGACTGTCCAGTCAGCAACTATGGGTAACGGTGGGG CTGATGGCTCCGTCAAAGATGCTGCTTCCGTTTCCAAGCCTTGATCAACGGCGTCGGCCG ACTTTATGATGAGGCTCGTCCAAAGTCTACGAAAATAAGGCGTCACTAGATGTTGGGACC TTCACGACGTCGTATTTGTGTAGACTTTGTGACTGAATCTAACAAACAACAAAAAAGCAA GAAAGAAGCCAATCCCAAGA |
STS Forward / Reverse: Reverse read
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Frying dragon/HyuuganatsuComment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): A255(Okitsu46gou)/G434(Kankitsu Chuukanbohon Nou5)
Comment: PCR_condition: 58C aneal
Figure legend:
 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Hae3Rank as CAPS marker: no homozygous cultivar with minor allele
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Experiments: CAPS pattern of Citrus collection
Rsetriction enzyme: Hae3Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
Rsetriction enzyme: Hha1
Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
Rsetriction enzyme: Hinc2
Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
Experiments: Segregation pattern
Population name: Jutaro unshu x Trovita orangeRestriction enzyme: Hae3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Hae3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Hae3
Comment: PCR_condition: 58C aneal
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Okitsu41gou
Restriction enzyme: Hae3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
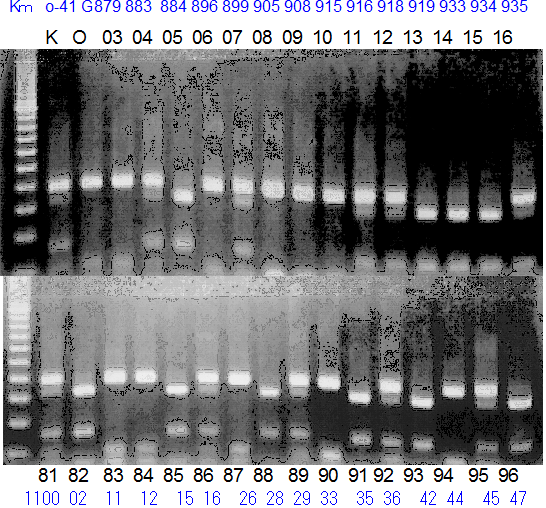 |
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese Acidless
Restriction enzyme: Hinc2
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID:Scaffold:
Start:
End:
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00305:mRNA_10.1Scaffold: C_unshiu_00305
Start: 69938
End: 73026
Gene function: Basic helix-loop-helix transcription factor
AGI map info (Shimada et al. 2014)
AGI LG: AGI-01AGI cM: 37.357
KmMg map info (Omura et al. 2003)
KmMg LG:KmMg cM:
JtTr map info (Omura et al. 2003)
JtTr LG: JtR-01SJtTr cM: 50
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1:DNA sequence around SNP1:
DNA sequence around SNP2:
DNA sequence around SNP3:
DNA sequence around SNP4: