Locus name: Al0304
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 54Main product (bp): 450
Comment on PCR:
F-primer: CGCAGATACCAGACACCAG
R-primer: GAGAAACCTCCGCATAATGAT
EST info
EST Accession number 1: C95332EST Accession number 2:
sequence-tagged site (STS seq)
STS:| CCCCAAAACCAGTATAAAGTGTGGCACATGCCCTTGTGTGACCCATGTGGTCAGCTCCCC CCTCCACCACCGCCTCCGCCGCCACCACCATCGCCGCCAAAGACTACATACTGTCCTCCG CAGGTGCAGACACCGCCGCCACCCAGGTTCATCTATGTGACTGGTGAGCCTGGGGAAATT TACCACACTGATTCTGATGACTGGGGGTTTTACTCGACTGCTCAACCAAATGTGGCTAAA AATCGGTTGCTTTTGGCTGCATGTGGTGTGTTGGGGCTCTTCATAATTTGGTAAAATTAA TGTAATATTGTGATCAAACATCTTAGATCTTGACAGTGTCAGTATCCTACATGCTACATA TACTTGTTAATTTCTCTCTTCATTCGAATTGATCATGTTTTATTTTGATAAGTGGACAAA ATCATTA |
STS Forward / Reverse: Foward read
STS:
| CCACCACCATCGCCGCCAAAGACTACATACTGTCCTCCGCAGGTGCAGACACCGCCGCCA CCCAGGTTCATCTATGTGACTGGTGAGCCTGGGGAAATTTACCACACTGATTCTGATGAC TGGGGGTTTTACTCGACTGCTCAACCAAATGTGGCTAAAAATTGGTTGCTTTTGGCTGCA TGTGGTGTGTTGGGGCTCTTCATAATTTGGTAAAATTAATGTAATATTGTGATCAAATAT CTTAGATCTTGACAGTGTCAGTATCCTACATGCTACATGTACTTGTTAATTTCTCTCTTC ATTCGAATTGATCATATTTTATTTTGATAAGTGGACAAAATCATT |
STS Forward / Reverse: Foward read
STS:
| GTGTGACCCATGTGGTCAGCTCCCCCCTCCGCCACCACCGCCTCCGCCGCCGCCNCCACC ACCATCGCCGCCAAAGACTACATACTGTCCTCCGCAGGTGCAGACACCGCCGCCACCCAG GTTCATCTATGTGACTGGTGAGCCTGGGGAAATTTACCACACTGATTCTGATGACTGGGG GTTTTACTCGACTGCTCAACCAAATGTGGCTAAAAATCGGTTGCTTTTGGCTGCATGTGG TGTGTTGGGGCTCTTCATAATTTGGTAAAATTAATGTAATATTGTGATCAAACATCTTAG ATCTTGACAGTGTCAGTATCCTACATGCTACATATACTTGTTAATTTCTCTCTTCATTCG AATTGATCATGTTTTATTTTGATAAGTGGACAAAATCATTANGGGGNAGGTTTCTCAA |
STS Forward / Reverse:
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-waseComment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Rsa1Rank as CAPS marker: Disagree by Marco analysis
Comment:
Figure legend:
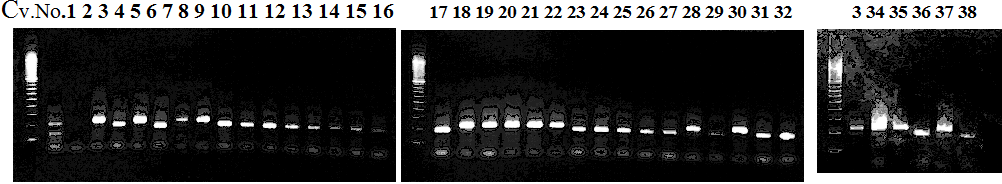
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Experiments: CAPS pattern of Citrus collection
Experiments: Segregation pattern
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese AcidlessRestriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Rsa1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Rsa1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
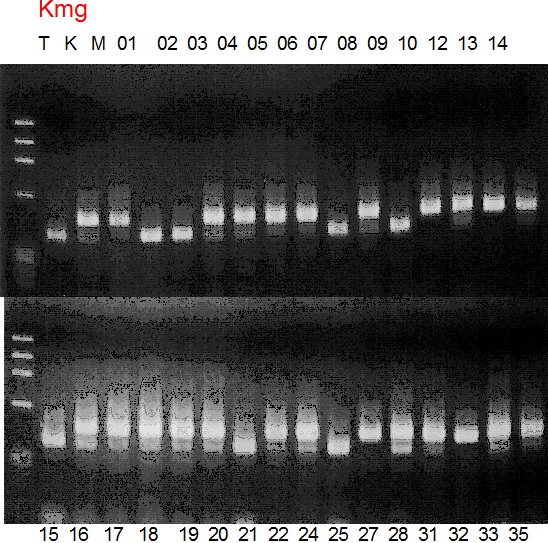
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Rsa1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Okitsu41gou
Restriction enzyme: Rsa1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10013536mScaffold: scaffold_6
Start: 22616979
End: 22619271
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00038:mRNA_27.1Scaffold: C_unshiu_00038
Start: 174815
End: 175255
Gene function: Nutrient reservoir, putative
AGI map info (Shimada et al. 2014)
AGI LG: AGI-04AGI cM: 46.361
KmMg map info (Omura et al. 2003)
KmMg LG: KmG-04KmMg cM: 16.744
JtTr map info (Omura et al. 2003)
JtTr LG:JtTr cM:
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG: NAT-04Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG: RP-04Rp/Sa cM: 54.617
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI259DNA sequence around SNP1:
| TACTTGTTAATTTCTCTCTTCATTCGAATTGATCAT[A/G]TTTTATTTTGATAAGTGGA CAAAATCATTANGGGGNAGGTTTCTCAA |
DNA sequence around SNP2:
| CACACTGATTCTGATGACTGGGGGTTTTACTCGACTGCTCAACCAAATGTGGCTAAAAAT [T/C]GGTTGCTTTTGGCTGCATGTGGTGTGTTGGGG |
DNA sequence around SNP3:
DNA sequence around SNP4: