Locus name: Al0326
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 60Main product (bp): 600
Comment on PCR:
F-primer: CAGAACAAAGCGATGGAACC
R-primer: CCCAAGTCCTGACCAACACTA
EST info
EST Accession number 1: C95360EST Accession number 2:
sequence-tagged site (STS seq)
STS:| AGGACCAAGCTGCCTCCGCAAAAGCCGCTATGGAGAAATCTCTCAATTTGAAGTCACCGA AGCAAATNCATCAGACGCCAGAATTGGAGACTGCCAAGGTGGCTGCTGAGCCCTTTGTGG GTTCCTCGTCGTCTGCTGCGGCCTCTCAACAGTTGAGTGTTGATCAGCCTGAGCCCCAAG CCAAGGGGCCTACCAGGTGCTTGAGTTGCAACAAGAAGGTCGGATTGACAGGGTTCAAGT GTAAGTGCGGGAGTACTTTCTGTGGAATCCATCGGTACCCAGAGAAGCATGATTGTACAT TTGATTTTAAGGTTACTGGTCGAGATGCTATTGCCAAAGCAAATCCTGTTGTTAAAGCTG ATAAGCTGGACAGGATCTGAAATCAGAGTCTGGTAATTTGGTTTGCTTTGGTATGGTAAT TGTCGTCGGTTGGGTTATTGTTGATTTAGCTTAGCTT |
STS Forward / Reverse: Foward read
STS:
| TGCCTCCGCAAAAGCCGCTATGGAGAAATCTCTCAATTTGAAGTCACCGAAGCAAATCCA TCAGACGCCAGAATTGGAGACTGCCAAGGTGGCTGCTGAGCCCTTTGTGGGTTCCTCGTC GTCTGCTGCGGCCTCTCAACAGTTGAGTGTTGATCAGCCTGAGCCCCAAGCCAAGGGGCC TACCAGGTGCTTGAGTTGCAACAAGAAGGTCGGATTGACAGGGTTCAAGTGTAAGTGCGG GAGTACTTTCTGTGGAATCCATCGGTACCCAGAGAAGCATGATTGTACATTTGATTTTAA GGTTACTGGTCGA |
STS Forward / Reverse: Foward read
STS:
| GGGGGACCAGCTACACCACATCTGAGCCAAAGCTCTGCGTCAACGGTTGTGGCTTCTTCG GCACTGCGGCGAATATGGGCTTATGCTCAAAGTGTTACAGGGATTTGCGGGTTAAGGAGG ACCAAGCTGCCTCCGCAAAAGCCGCTATGGAGAAATCTCTCAATTTGAAGTCACCGAAGC AAATCCATCAGACGCCAGAATTGGAGACTGCCAAGGTGGCTGCTGAGCCCTTTGTGGGTT CCTCGTCGTCTGCTGCGGCCTCTCAACAGTTGAGTGTTGATCAGCCTGAGCCCCAAGCCA AGGGGCCTACCAGGTGCTTGAGTTGCAACAAGAAGGTCGGATTGACAGGGTTCAAGTGTA AGTGCGGGAGTACTTTCTGTGGAATCCATCGGTACCCAGAGAAGCATGATTGTACATTTG ATTTTAAGGTTACTGGTCGAGATGCTATTGCCAAAGCAAATCCTGTTGTTAAAGCTGATA AGCTGGACAGGATCTGAAATCAGAGTCTGGTAATTTGGTTTGCTTTGGTATGGTAATTGT CGTCGGTTGGGTTATTGTT |
STS Forward / Reverse: Reverse read
STS:
| TCTGATTTCAGATCCTGTCCAGCTTATCAGCTTTAACAACAGGATTTGCTTTGGCAATAG CATCTCGACCAGTAACCTTAAAATCAAATGTACAATCATGCTTCTCTGGGTACCGATGGA TTCCACAGAAAGTACTCCCGCACTTACACTTGAACCCTGTCAATCCGACCTTCTTGTTGC AACTCAAGCACCTGGTAGGCCCCTTGGCTTGGGGCTCAGGCTGATCAACACTCAACTGTT GAGAGGCCGCAGCAGACGACGAGGAACCCACAAAGGGCTCAGCAGCCACCTTGGCAGTCT CCAATTCTGGCGTCTGATGGATTTGCTTCGGTGACTTCAAATTGAGAGATTTCTCCATAG CGGCTTTTGCGGAGGCAGCTTGGTCCTCCTTAACCCGCAAATCCCTGTAACACTTTGAGC ATAAGCCCATATTCGCCGCAGTGCCGAAGAAGCCACAACC |
STS Forward / Reverse: Reverse read
STS:
| TGCCTCCGCA AAAGCCGCTA TGGAGAAATC TCTCAATTTG AAGTCACCGA AGCAA ATNCA TCAGACGCCA GAATTGGAGA CTGCCAAGGT GGCTGCTGAG CCCTTTGTGG GTTCCTCGTGGTCTGCTGCG GCCTCTCAAC AGTTGAGTGT TGATCAGCCT GAGCC CCAAG CCAAGGGGCCTACCAGGTGC TTGAGTTGCA ACAAGAAGGT CGGATTGACA GGGTTCAAGT |
STS Forward / Reverse:
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-waseComment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Nde2Rank as CAPS marker: Available marker
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Experiments: CAPS pattern of Citrus collection
Experiments: Segregation pattern
Population name: Jutaro unshu x Trovita orangeRestriction enzyme: Mbo1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
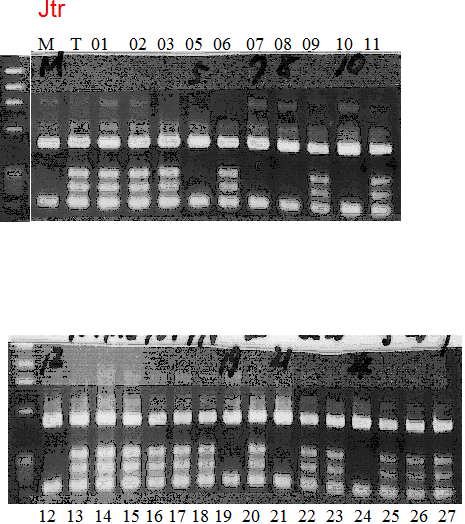 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Mbo1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
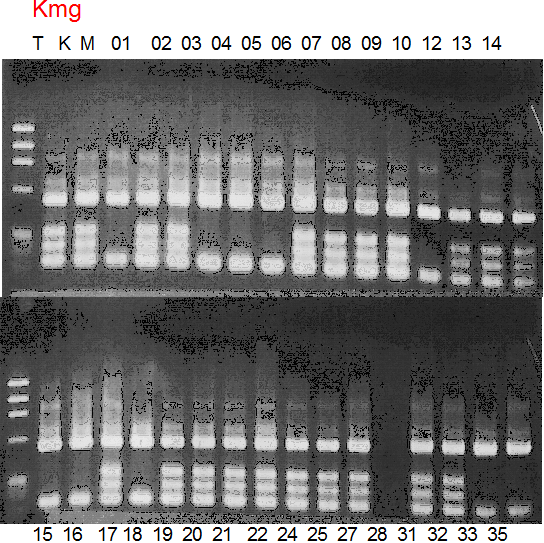 |
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese Acidless
Restriction enzyme: Nde2
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Nde2
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
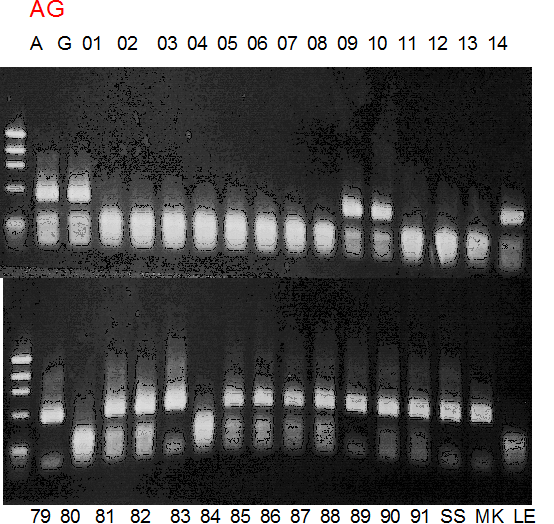 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10022558mScaffold: scaffold_3
Start: 6409311
End: 6411254
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00443:mRNA_7.1Scaffold: C_unshiu_00443
Start: 62532
End: 64193
Gene function: Zinc finger A20 and AN1 domain-containing stress-associated protein 8
AGI map info (Shimada et al. 2014)
AGI LG: AGI-05AGI cM: 42.609
KmMg map info (Omura et al. 2003)
KmMg LG: KmG-05SKmMg cM: 32.841
JtTr map info (Omura et al. 2003)
JtTr LG: JtR-05JtTr cM: 16.025
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG: RP-05Rp/Sa cM: 27.235
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K: KSNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI065DNA sequence around SNP1:
| GTCTGCTGCGGCCTCTCAACAGTTGAGTGTTGA[T/G]CAGCCTGAGCCCCAAGCCAAGG GGCCTACCAGGTGCTTGAGTTGCAACAAGAAGGTCGGA |
DNA sequence around SNP2:
| TGCCTCCGCAAAAGCCGCTATGGAGAAATCTCTCAATTTGAAGTCACCGAAGCAAAT[A/ C]CATCAGACGCCAGAATTGGAGACTGCCAAGGTGGCTGCTGAGCCCTTTGTGGGTTCCT CG |
DNA sequence around SNP3:
DNA sequence around SNP4: