Locus name: Al0413
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 56Main product (bp): 1100
Comment on PCR:
F-primer: ATACCATTCCGGTCCTGAAAG
R-primer: GCTCCACTTGCTCCAAACA
EST info
EST Accession number 1: C95392EST Accession number 2: DC892983
sequence-tagged site (STS seq)
STS:| CGTGCCTTCAAAGACAAGTTCAAGGACACTTCTTGGTGACACCATTGTGGTAAATACTCA CATAACTTGTTAATACTTTTCAATTTAGATTGTTAAAGTAGAAGGCTGGTTGATGAATGC TAAATTATTTGTTACATATAGGAAATCCAAGTGGGAATGGGTCCAGCTGGTGAGCTGCGT TACCCTTCGTATCCAGAGCAGAATGGGACATGGAAATTCCCAGGAATTGGAGCTTTCCAG TGTTATGATAAGGTATTTGCTAATTTTTTTTTTCTTGNGTTGTTTTGAGTCTGGTAAATT TGCGGNGGGATTTGTAGCGNGTGGTTCTCATATTGG |
STS Forward / Reverse: Foward read
STS:
| GTTTGGAGTTCTGGTAAATTTGCGGTGGGATTNGTAGCGTGTGGTTCTCATATTGGTGTT TGATGTCTTGATGCAGTATATGCTCAGTAGCTTAAAAGCAGCAGCTGAATCTGCTGGTAA GCCAGAATGGGGCAGCACAGGCCCTACTGATGCTGGACATTACAACAACTGGCCAGAAGA TACACAGTTTTTCCGAAAAGAAAATGGTGGTTGGTGTAGCCCTTATGGTGAATTTTTCCT AAGCTGGTATTCGCAGATGCTCTTAGATCACGGAGAGAGAATTCTCTCATCTGCCAAGGC AATTTTCGATGCTACAGGTGTTAAGATCTCGGTTAAGGTTGCAGGAATCCACTGGCACTA TGGGTCTAGGTCCCATGCCCCTGAGCTCACAGCTGGATATTACAATACACGATTCCGTGA TGGTTACCTCCCTATTGCCCAGATGCTAGCACGACATGGTGCCATATTCAACTTTACTTG CATAGAGATGCGTGATCATGAACAACCACAAGATGCCCTTTGTGCACCTGAGAAGCTAGT CAAGCAAGTGGCTTCAGCAACTCAGAAAGCACACGTTCCACTGGCTGGGGAAAATGCGTT GCCACGTTATGATGAATACGCTCACGAGCAGATCTTGCGAGCAGCATCACTTGATGTAGA CAAACAGATGTGTGCATTCACGTACTTGAGGATGAATCCACATTTATTCCAACCCGATAA TTGGAGACAGTTTGTGGC |
STS Forward / Reverse: Reverse read
STS:
| AATAAATGTGGATTCATCCTCAAGTACGTGAATGCACACATCTGTTTGTCTACATCAAGT GATGCTGCTCGCAAGATCTGCTCGTGAGCGTATTCATCATAACGTGGCAACGCATTTTCC CCAGCCAGTGGAACGTGTGCTTTCTGAGTTGCTGAAGCCACTTGCTTGACTAGCTTCTCA GGTGCACAAAGGGCATCTTGTGGTTGTTCATGATCACGCATCTCTATGCAAGTAAAGTTG AATATGGCACCATGTCGTGCTAGCATCTGGGCAATAGGGAGGTAACCATCACGGAATCGT GTATTGTAATATCCAGCTGTGAGCTCAGGGGCATGGGACCTAGACCCATAGTGCCAGTGG ATTCCTGCAACCTTAACCGAGATCTTAACACCTGTAGCATCGAAAATTGCCTTGGCAGAT GAGAGAATTCTCTCTCCGTGATCTAAGAGCATCTGCGAATACCAGCTTAG |
STS Forward / Reverse: Reverse read
STS:
| AATTTTCGAT GCTACAGGTG TTAAGATCTC GGTTAAGATT GCAGGAATCC ACTGG CACTA TGGGTCTAGG TCCCATGCCC CTGAGCTCAC AGCTGGATAT TACAATACAC GATTCCGTGA TGGTTACCTC CCTATTGCCC AGATGCTAGC ACGACATGGT GCCA TATTCA ACTTTACTTG CATAGAGATG CGTGATCATG AACAACCACA AGATGCCCT T TGTGCACCT |
STS Forward / Reverse:
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-waseComment:
Figure legend:
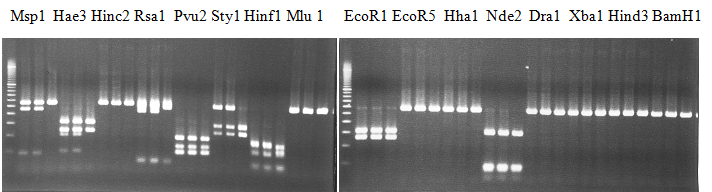 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Msp1Rank as CAPS marker: Available marker
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Experiments: CAPS pattern of Citrus collection
Experiments: Segregation pattern
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese AcidlessRestriction enzyme: Msp1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Msp1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Msp1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10004620mScaffold: scaffold_9
Start: 7579900
End: 7583093
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00063:mRNA_61.1Scaffold: C_unshiu_00063
Start: 483279
End: 486427
Gene function: Chloroplast beta-amylase 3
AGI map info (Shimada et al. 2014)
AGI LG: AGI-03AGI cM: 46.197
KmMg map info (Omura et al. 2003)
KmMg LG: KmG-032KmMg cM: 19.57
JtTr map info (Omura et al. 2003)
JtTr LG: JtR-031JtTr cM: 10.395
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG: RP-03Rp/Sa cM: 33.559
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K: KSNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI275DNA sequence around SNP1:
| TGGATATTACAATACACGATTCCGTGATGGTTACCTCCCTATTGCCCAGATGCTAGCACG [A/G]CATGGTGCCATATTCAACTTTACTTGCATAGAGATGCGTGATCATGAACAACCAC AAGAT |
DNA sequence around SNP2:
| ATCTTGTGGTTGTTCATGATCACGCATCTCTATGCAAGTAAAGTTGAATATGGCACCATG [T/C]CGTGCTAGCATCTGGGCAATAGGGAGGTAACCATCACGGAATCGTGTATTGTAAT ATCCA |
DNA sequence around SNP3:
DNA sequence around SNP4: