Locus name: Al0415
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 56Main product (bp): 2400
Comment on PCR:
F-primer: TCCATCTCCCGAAGCCATTG
R-primer: AATTGCCCTGCCTTTTCCAGA
EST info
EST Accession number 1: C95396EST Accession number 2: DC892985
sequence-tagged site (STS seq)
STS:| NGTAATTTTTCAAGNCTGCACGTTTATAATATACTGGCTGCTAAATGGTTTTTGATGTAG CAGGTGCCGCAAGACCTGGGAAAGTTGTCAGTTCAGTGTTGCGTTACAACAATTGTGGAG CAGCAGCAGCTGCAGAGGCTCTTGATCAGCGGAGAGTGGTTAGGAATCCTGCTGTTTCAA CTCAATATGCTGCCAGTTCATATCCAAGAAGAAGCTCAGCATGTAAAAACGAGAGGGTGA ATGATGAATCTGTTGAAGGTTCCAATGGATTGCAGCCAAAGCCTCAGTACATAGCAAGGA AAGTTGCTGCTGCTCAAGGTGGTTCTGGAAGCAACTGGTATTGATTCATTGCTATGTCGT TCCTACAGTGGACTCAACCGTCTACACTCCGCTTGATGATGCTAGTTACACAGCATCCAA GATTGATCTGCTCGCAAGTGGTAAATATCAGACACGGTATGTTTTCCTGGTGCTACATAC NATCTCAGAGTTTAAAGGATTAGGAGTTTGATATTTTATGTTGGAGAAAGCCCTAAAAGT TGTAATTTATAGCACTAACTGAAAACGTAGGAGGGGTTAGTTCGGGTTCAACTGTCCACG ACCCTCTTCC |
STS Forward / Reverse: Reverse read
STS:
| CCGAACTAACCCCTCCTACGTTTTCAGTTAGTGCTATAAATTACAACTTTTAGGGCTTTC TCCAACATAAAATATCAAACTCCTAATCCTTTAAACTCTGAGATGGTATGTAGCACCAGG AAAACATACCGTGTCTGATATTTACCACTTGCGAGCAGATCAATCTTGGATGATGTGTAA CTAGCATCATCAAGCGGAGTGTAGACGGTTGAGTCCACTGTAGGAACGACATAGCAATGA ATCAATACCAGTTGCTTCCAGAACCACCTTGAGCAGCAGCAACTTTCCTTGCTATGTACT GAGGCTTTGGCTGCAATCCATTGGAACCTTCAACAGATTCATCATTCACCCTCTCGTTTT TACATGCTGAGCTTCTTCTTGGATATGAACTGGCAGCATATTGAGTTGAAACAGCAGGAT TCCTAACCACTCTCCGCTGATCAAGAGCCTCTGCAGCTGCTGCTGCTCCACAATTGTTGT AACGCAACAC |
STS Forward / Reverse: Reverse read
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-waseComment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: EcoR1Rank as CAPS marker: Available marker
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
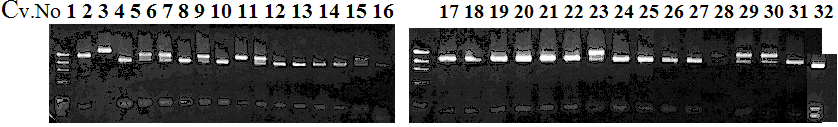 |
Experiments: CAPS pattern of Citrus collection
Experiments: Segregation pattern
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)Restriction enzyme: EcoR1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: EcoR1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: EcoR1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Okitsu41gou
Restriction enzyme: EcoR1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10004660mScaffold: scaffold_9
Start: 2732900
End: 2738896
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00040:mRNA_76.1Scaffold: C_unshiu_00040
Start: 632823
End: 638463
Gene function: Mitogen-activated protein kinase 14
AGI map info (Shimada et al. 2014)
AGI LG: AGI-03AGI cM: 24.885
KmMg map info (Omura et al. 2003)
KmMg LG: KmG-031KmMg cM:
JtTr map info (Omura et al. 2003)
JtTr LG:JtTr cM:
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG: NAT-03Km/O41 cM: 41.576
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI120DNA sequence around SNP1:
| AACTTTTAGGGCTTTCTCCAACATAAAATATCAAACTCCTAATCCTTTAAACTCTGAGAT [A/G]GTATGTAGCACCAGGAAAACATACCGTGTCTGATATTTACCACTTG |
DNA sequence around SNP2:
| TGTGTAACTAGCATCATCAAGCGGAG[A/T]GTAGACGGTTGAGTCCACTGTAGGAAC |
DNA sequence around SNP3:
DNA sequence around SNP4: