Locus name: Bf0115
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 58Main product (bp): 3000
Comment on PCR:
F-primer: TTAATGAGATACCGAAGTGTC
R-primer: GCATTGCCTATAAGAAGAATA
EST info
EST Accession number 1: DC885474EST Accession number 2:
sequence-tagged site (STS seq)
STS:| CACATCTATTTTCCTTTCAATTTCCCTTTAAATCTCCTGGAGGGTTTATGATTTTGAATA TCTCGTAAGGGCATAATCCCATTAATTGTTGGTTTTTCTTTTTCTAATTGGCATTTTTGA AATTTTGCCCCTCAAGGACTTAAAATATGAGGAAAACTCTGACAAAGCAGTACAGTGCTT GAATGATATGGTCACCAATGCTCTGATGCATGTGGAAGATTGTTTGAAGTACATGTCTGC TTTAAGGGATCATGCTATATTCCGATTCTGTGCTATCCCTCAGGTGCGCTGCTTCTAGTA TATTCTGATTTGAGAATTAATCTTGGAATTTTGGTATCAACATTTATTTTTAACTGTTAC AATACCATCCACCCCAAATTAAACTTACCCCCCTCGAAGATTGGTGTGAGATGGATGAAT CCATGACCTCTTGCTTCCACTGGAAGAGGCCAAACCAATT |
STS Forward / Reverse:
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): A255(Okitsu46gou)/G434(Kankitsu Chuukanbohon Nou5)Comment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
 |
Experiments: CAPS pattern of hybrid cultivar
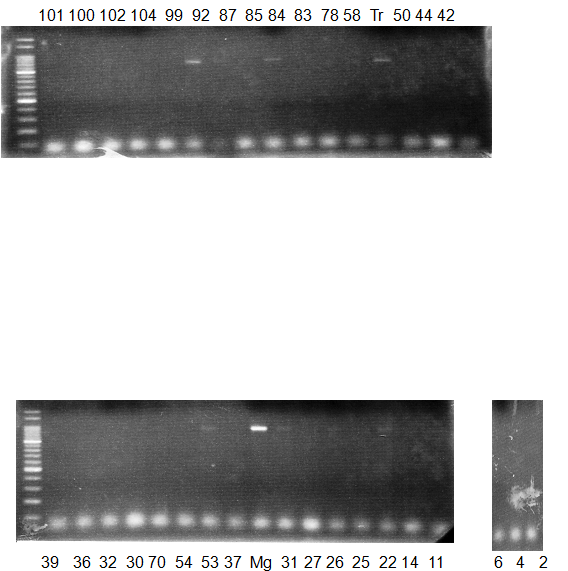
Experiments: CAPS pattern of Citrus collection
Rsetriction enzyme: NCComment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
Experiments: Segregation pattern
Population name: Kiyomi x Okitsu41gouRestriction enzyme: Hind3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Hind3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese Acidless
Restriction enzyme: Msp1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10029066mScaffold: scaffold_8
Start: 6317205
End: 6322458
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00484:mRNA_11.3Scaffold: C_unshiu_00484
Start: 48216
End: 54250
Gene function: Squalene synthase
AGI map info (Shimada et al. 2014)
AGI LG: AGI-08AGI cM: 58.587
KmMg map info (Omura et al. 2003)
KmMg LG:KmMg cM:
JtTr map info (Omura et al. 2003)
JtTr LG:JtTr cM:
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG: NAT-081Km/O41 cM: 17.717
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping:Cultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI054DNA sequence around SNP1:
| GCTTTAAGGGATCATGCTATATTCCGATTCTGTGCTATCCCTCAGGTGCGCTGCTTCTAG [T/C]ATATTCTGATTTGAGAATTAATCTTGGAATTTTGGTATCAACATTTATTTTTAAC TGTTA |
DNA sequence around SNP2:
| CTCGTAAGGGCATAATCCCATTAATTGTTGGTTTTTCTTTTTCTAATTGGCATTTTTGAA [A/T]TTTTGCCCCTCAAGGACTTAAAATATGAGGAAAACTCTGACAAAGCAGTACAGTG CTTGA |
DNA sequence around SNP3:
DNA sequence around SNP4: