Locus name: Bf0130
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 56Main product (bp): 600
Comment on PCR:
F-primer: GCCTGGAATCCGTGAT
R-primer: GACCCTAATCTATTGCCAAGT
EST info
EST Accession number 1: DC885666EST Accession number 2:
sequence-tagged site (STS seq)
STS:| GCACTTTCAACACTGTTCTCTGTCCTTAAAGAGCTGAAATGGAGACCGGACACCAAATCT CTTCTTGCGTCAAGTCTTGCGGGGACTGGCATGGGGAAGAGAGGTGGTAAATTTTTCAAG AAGATGTGAGGCCTGAATTAGTCTCTTAACAAGATTAGATTTTGCACGTTTTT-TCTCAT GACCAGTTGACCACGATCCCAGCCACTGTCCAAATGGTCTTTCTGTGCGAAATTCTCTTC ATCAAATATAATCTGATTCCTTGTCCATTTTTAGCCAGTTAACTTGTATTTTGACAAAGG CAGATTGTTGTTTAGTCTCATCCTTTCTGGTATGAGTGTGGAATACCAGCCGAGGGATGA AAGAGTATGATAAATTTGCAGATCTCCTTTCTTGATGCACATCAGGCCGAAGACGCCTTT GCTCATTTCAATATCAAGTGCAAACATCCGAGAAGATGTCTCGGTCTTAGA |
STS Forward / Reverse:
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): A255(Okitsu46gou)/G434(Kankitsu Chuukanbohon Nou5)Comment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
 |
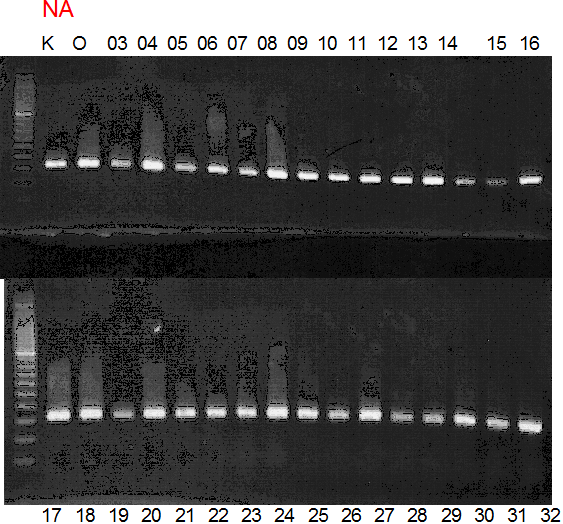
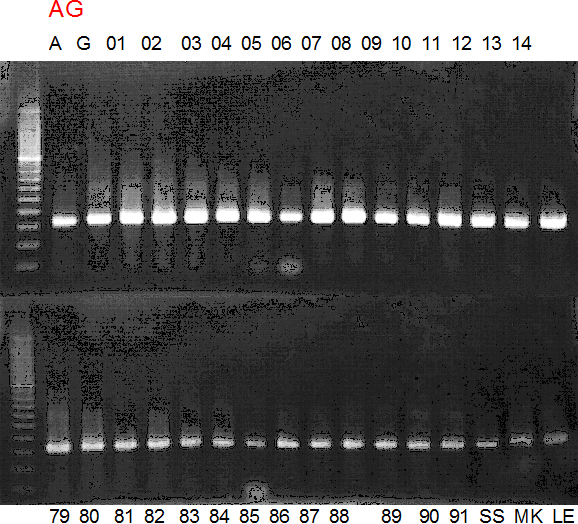
Experiments: CAPS pattern of hybrid cultivar
Experiments: CAPS pattern of Citrus collection
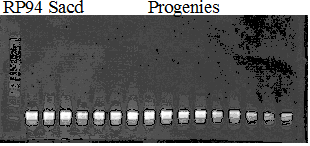
Experiments: Segregation pattern
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese AcidlessRestriction enzyme: Hae3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Okitsu41gou
Restriction enzyme: Pvu2
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Pvu2
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10004858mScaffold: scaffold_9
Start: 26981498
End: 26984277
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00095:mRNA_24.1Scaffold: C_unshiu_00095
Start: 228406
End: 230902
Gene function: F-box family protein isoform 1
AGI map info (Shimada et al. 2014)
AGI LG: AGI-03AGI cM: 40.452
KmMg map info (Omura et al. 2003)
KmMg LG:KmMg cM:
JtTr map info (Omura et al. 2003)
JtTr LG:JtTr cM:
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping:Cultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI063DNA sequence around SNP1:
| TCTCATGACCAGTTGACCACGATCCCA[A/G]CCACTGTCCAAATGGTCTTTCTGTGCGA AATTCTCTTCATCAAATATAATCTGATTCCTT |
DNA sequence around SNP2:
| CAAAGGCAGATTGTTGTTTAGTCTCATCCTTTCTGGTATGAGTGTGGAATACCAGCCGAG [A/G]GATGAAAGAGTATGATAAATTTGCAGATCTCCTTTCTTGATGCACATCAGGCCGA AGACG |
DNA sequence around SNP3:
DNA sequence around SNP4: