Locus name: Bf0200
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 58Main product (bp): 1600
Comment on PCR:
F-primer: AAAACAGCACCTCGCACAAT
R-primer: GTGACTGTCCGGTTGAAAACA
EST info
EST Accession number 1: DC886299EST Accession number 2: DC886300
sequence-tagged site (STS seq)
STS:| TAACATGGAATCTGGCACAATTATGCATCTTGACGCCAAAAGATGGCCTGATGCTCAAGC AGATGATGCATCAGCTTTCTACAAGGTGATTTTTGAATAAAAAGCTAAATTCCAATCTTC TATTTCAAGAGGGGAAAATGATTTTTAAAATTTATCTAAGTCTGCATGATTTGGCTGCTG CATTTAAGAATTTGAGCTCAAATTTTCTTTTGGTGACATGTGAAAAGAAGTTTCTGGTTC ATGCTAACCGGACAGCAGTTTGGTTGAATTTTGTATTGATTAGAATATTTATCTACGGTT GTATTTCACTCTTCAGTTCTGTTAGAGATTTATGAATATCATTAACAAGACTTGCTTCAA GCTTGTTGTGAGCATCTTATCTTCATTTGAATTGGCTGCACATTTGATATTTCAATCATA ACTGATCTT |
STS Forward / Reverse:
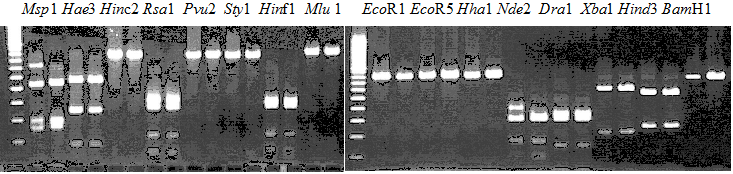
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): A255(Okitsu46gou)/G434(Kankitsu Chuukanbohon Nou5)Comment:
Figure legend:
 |
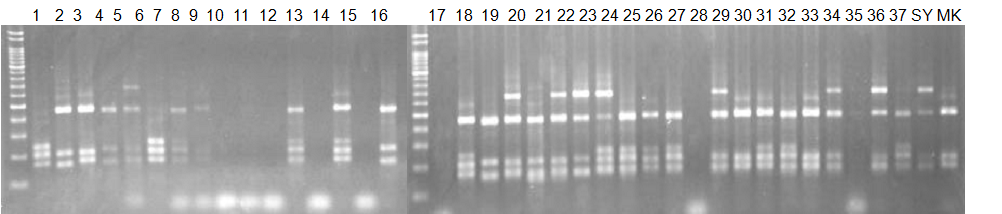
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Msp1Rank as CAPS marker: Others
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Experiments: CAPS pattern of Citrus collection
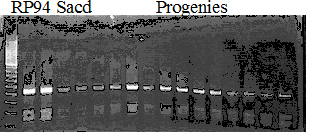
Experiments: Segregation pattern
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese AcidlessRestriction enzyme: Msp1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
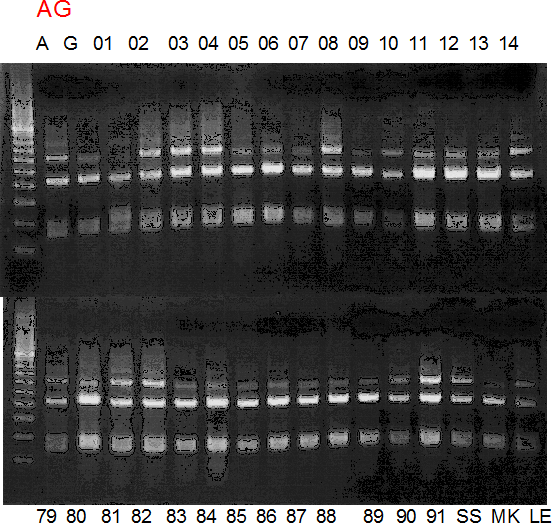
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Msp1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10014367mScaffold: scaffold_2
Start: 29022433
End: 29030724
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00022:mRNA_44.1Scaffold: C_unshiu_00022
Start: 308324
End: 316293
Gene function: Glutamine-tRNA ligase / glutaminyl-tRNA synthetas, putative isoform 1
AGI map info (Shimada et al. 2014)
AGI LG:AGI cM:
KmMg map info (Omura et al. 2003)
KmMg LG:KmMg cM:
JtTr map info (Omura et al. 2003)
JtTr LG:JtTr cM:
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping:Cultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI335DNA sequence around SNP1:
| AAAATTTATCTAAGTCTGCATGATTTGGCTGCTGCATTTAAGAATTTGAGCTCAAATTTT [C/G]TTTTGGTGACATGTGAAAAGAAGTTTCTGGTTCATGCTAACC |
DNA sequence around SNP2:
DNA sequence around SNP3:
DNA sequence around SNP4: