Locus name: Fb0110
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 60Main product (bp): 2400
Comment on PCR:
F-primer: CACGAACTGTGCTTGTCA
R-primer: CCTTAAAATTTGCCTCCTAAT
EST info
EST Accession number 1: DC888023EST Accession number 2: DC888024
sequence-tagged site (STS seq)
STS:| TGTGTATTTATGTGCAAATAAAAGTACTCAGTGTGATGTTGATTTCATTTTTACTTGCTT TTGCAGAGAGAAAGATGCTGACACAAGGGGCTTTCTCGAAGCAGCGAAAGCATGTCTTCA AATTTGTTGCGTCCATGGAGTACCTCTGCTGATTAATGACCGCATCGATATTGCCCTTGC TTGTGATGCTGATGGTGTGCATCTAGGTCAGTCTGACATGCCTGCTCGTACTGCCCGTGC TCTTCTTGGTCCTGATAAGATCATTGGCGTATCGTGCAAAACACCTGAAGAAGCCCATCA AGCTTGGATTGATGGTGCCAATTACATTGGTTGTGGTGGAGTATATCGAACTAACACAAA GGCAAATAATCTCACTGTCGGTTTGGATGGATTGAAAACTGTTTGTTTGGCTTCCAAGTT ACCTGTGGTTGCAATTGGTGGCATCGGCATTTCAAATGCATCTGACGTGATGAAAATTGG TGTATCAAATCTTAAAGGTGTTGCCGTTGTGTCAGCTCTTTTTGATAGGGAATGTATCTT G |
STS Forward / Reverse:
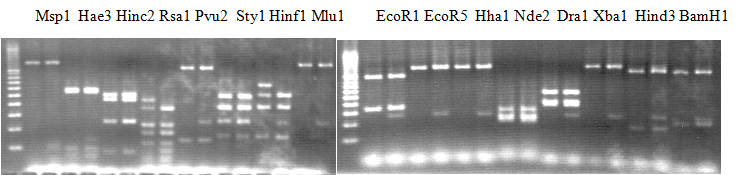
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): A255(Okitsu46gou)/G434(Kankitsu Chuukanbohon Nou5)Comment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-wase
Comment:
Figure legend:
 |
Experiments: CAPS pattern of hybrid cultivar
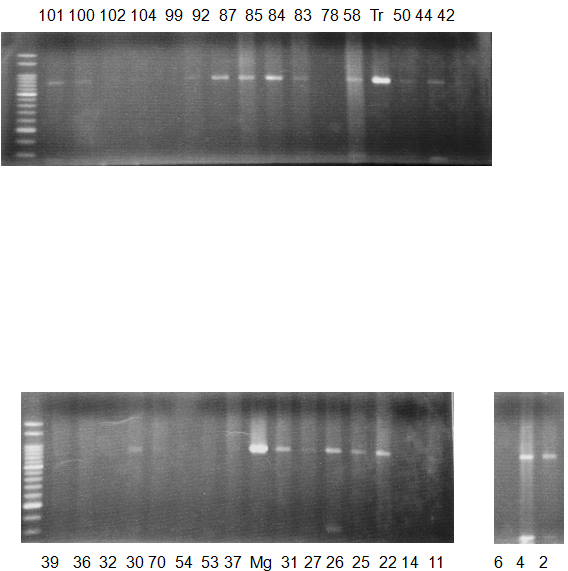
Experiments: CAPS pattern of Citrus collection
Rsetriction enzyme: NCComment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
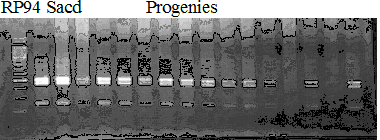
Experiments: Segregation pattern
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese AcidlessRestriction enzyme: Hinc2
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
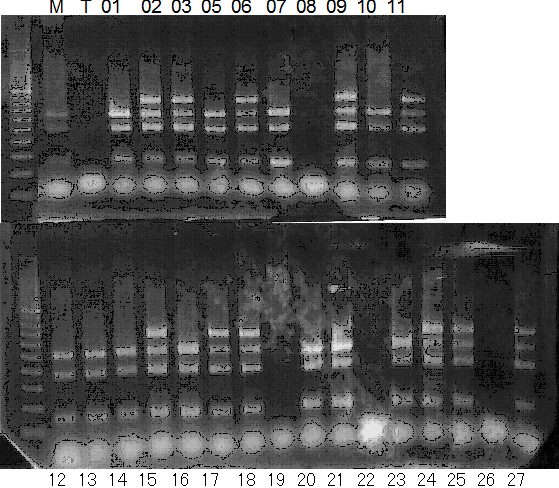
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10000900mScaffold: scaffold_5
Start: 8512380
End: 8517835
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00033:mRNA_4.1Scaffold: C_unshiu_00033
Start: 37755
End: 43368
Gene function: Bifunctional protein thiED
AGI map info (Shimada et al. 2014)
AGI LG: AGI-09AGI cM: 85.81
KmMg map info (Omura et al. 2003)
KmMg LG: KmG-09KmMg cM: 23.401
JtTr map info (Omura et al. 2003)
JtTr LG: JtR-09SJtTr cM: 5.362
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping:Cultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI246DNA sequence around SNP1:
| GAAAGATGCTGACACAAGGGGCTTTCTCGAAGCAGCGAAAGCATGTCTTCAAATTTGTTG [T/C]GTCCATGGAGTACCTCTGCTGATTAATGACCGCATCGATATTGCCCT |
DNA sequence around SNP2:
DNA sequence around SNP3:
DNA sequence around SNP4: