Locus name: Gn0067
CAPS PCR info
PCR Anneal-Temp (℃) / Method: TdMain product (bp): 550
Comment on PCR:
F-primer: AGACCTTATGGCAGAGTTAAT
R-primer: CGGAATGACACAATGCTC
EST info
EST Accession number 1: AB114652EST Accession number 2:
sequence-tagged site (STS seq)
STS:| GCAAAAATGCATAACCAATGGTGTTAAGTTTCACCAAGCTAAAGTTATTAAGGTTATTCA TGAAGAGTCCAAATCTTTGTTGATTTGCAATGATGGTGTGACAATTCAGGCTGCCGTGGT TCTTGATGCTACGGGGTTCTCTAGGTGTCTTGTCCAGTATGATAAGCCCTATAATCCAGG TTACCAAGTGGCATATGGAATACTAGCTGAGGTAGAAGAGCACCCGTTTGATTTAGACAA GATGGTTTTCATGGATTGGAGAGATTCGCATCTGAACAACAATTCGGAGCTCAAAGAGGC AAATAGCAAAATTCCTACTTTTCTTTATGCCATGCCCTTTTCGTCAAACAGGATATTTCT TGAAGAGACTTCGCTAGTGGCGCGGCCTGGAGTGCCAATGAAAGATATCCAGGAAAGAAT GGTGGCTAGA |
STS Forward / Reverse:
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): A255(Okitsu46gou)/G434(Kankitsu Chuukanbohon Nou5)Comment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
 |
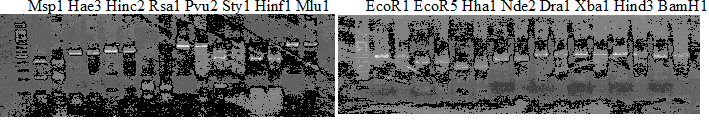
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Hae3Rank as CAPS marker: Others
Comment:
Figure legend: DNA sample: Lane 2'=Ethlog citron, 23'=Duncan GF, Hrd=Hirado buntan, HgNt = Hyuuganatsu
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Resriction enzyme: Hinf1
Rank as CAPS marker: Others
Comment:
Figure legend: DNA sample: Lane 2'=Ethlog citron, 23'=Duncan GF, Hrd=Hirado buntan, HgNt = Hyuuganatsu
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
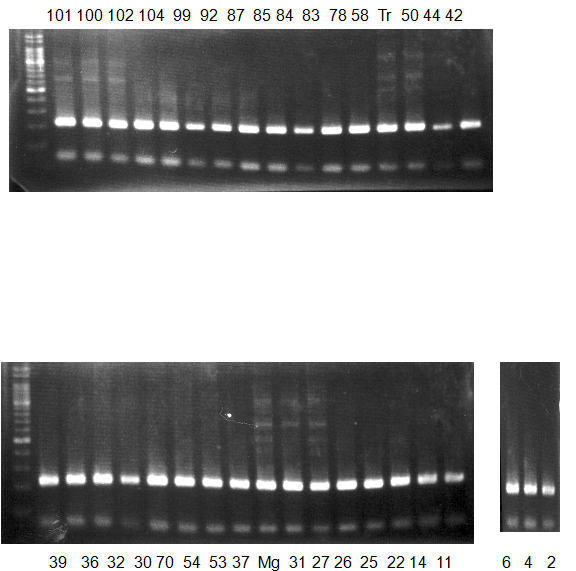
Experiments: CAPS pattern of Citrus collection
Rsetriction enzyme: Hae3Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
Experiments: Segregation pattern
Target on C. clementina (v1.0)
Transcript ID: Ciclev10004730mScaffold: scaffold_9
Start: 22728310
End: 22730086
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00295:mRNA_17.1Scaffold: C_unshiu_00295
Start: 207257
End: 208985
Gene function: Lycopene beta-cyclase
AGI map info (Shimada et al. 2014)
AGI LG: AGI-03AGI cM: 46.274
KmMg map info (Omura et al. 2003)
KmMg LG:KmMg cM:
JtTr map info (Omura et al. 2003)
JtTr LG:JtTr cM:
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping:Cultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI204DNA sequence around SNP1:
| GCAAAAATGCATAACCAATGGTGTTAAGTT[T/C]CACCAAGCTAAAGTTATTAAGGTTA TTCATGAAGAGTCCAAATCTTTGTTGATTTGCAAT |
DNA sequence around SNP2:
| CCCTATAATCCAGGTTACCAAGTGGCATATGGAATACTAGCTGAGGTAGAA[C/G]AGCA CCCGTTTGATTTAGACAAGATGGTTTTCATGGATTGGAGAGATTCGCATCTGAACA |
DNA sequence around SNP3:
DNA sequence around SNP4: