Locus name: If0003
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 62Main product (bp): 1600
Comment on PCR:
F-primer: GACAATCAATCACCCGTCGTT
R-primer: TTGAGGCAGGGGCTGAAT
EST info
EST Accession number 1: DC893584EST Accession number 2: DC893585
sequence-tagged site (STS seq)
STS:| ATGGAGCATGCCTATTCAGTACAAGGACTCAATTATGGAATCTACTTTGAATTGTCGGCA GAATGGTAGTCTCTTTGATGTCTCCCATATGTGTGGGCTGAGCCTCAAAGGGAAGGATTG CGTCCCTTTCCTTGAGAAGCTTGTGATCGCTGATGTTGCTGGACTTGCCCCAGGAACTGG GACTCTTACAGTCTTTACAAATGAAAATGGGGGTTCAATTGATGATTCAGTTATTACCAA GGTGAAGGATGATCACATATACCTTGTTGTGAATGCTGGCTGCAGGGATAAGGATCTTGC TCACATTGAGGCACATATGAAGAGTTTCACAGCCAAAGGTGGGGATGTCTCG |
STS Forward / Reverse: Foward read
STS:
| CTTTGCCTCAGATGCTGAAAACCTNAAAAAAACTGCCCTTCATGACTTTCATGTGGCTAA TGGTGGAAAGATGGTTCCATTTGCTGCGATGGAGCATGCCTATTCAGTACAAGGACTCAA TTATGGAATCTACTTTGAATTGTCGGCAGAATGGTAGTCTCTTTGATGTCTCCCATATGT GTGGGCTGAGCCTCAAAGGGAAGGATTGCGTCCCTTTCCTTGAGAAGCTTGTGATCGCTG ATGTTGCTGGACTTGCCCCAGGAACTGGGACTCTTACAGTCTTTACAAATGAAAATGGGG GTTCAATNGATGATTCAGTTATTACCAAGGTGAAGGATGATCACATATACCTTGTTGTGA ATGCTGGCTGCAGGGATAAGGATCTTGCTCACATTGAGGCACATATGAAGAGTTTCACAG CCAAAGGTGGGGATGTCTCGTGGCACATCCATGATGAGAGATCTCTTCTTGCCCTTCAGG TTACTCAGTGCACTGTTATAGTCTTTTACCTGGCCTTGCTTTTGTTTCACCCCTTGCTTG AGATGTGATGAATTTAGTTCAGTTTCTCAGAGTTAATATTCTTTATATTTCTAGATANGG TATCTGTGCTAGGTATGTGTGTATCTGAATTGATTTATCTCTTCANCAATTGTTTATTCC TATTTGAAGTCTTCATGAATTAGAATAGATCCGCAACAAATAATGGCCGTATGGCAGGCT CAAAGTTGATGATTANCGTTTCTTCACCG |
STS Forward / Reverse: Foward read
STS:
| TTTTTTGGTACCATATACATTTCAAGTTCTTCTGTCTTTATTATAAGTTGTATGAATAGG CTGTTCGGGATTTTTAACTACCTTTTTTTGTCTATTACAATTCTCAGGTACACTGGTGAA GATGGATTTGAAATCTCTGTTCCATCTGAGCGTGCAGTGGATCTTGCCAAAGCAATTCTG GAGAAATCTGAAGGAAAGGTGAGGTTGACAGGTNCTNCGGTGCTTNGAGGAGCACGTGCT GCCGACTTGACACGCTAGGCTTAATGGTTTATGAT |
STS Forward / Reverse: Reverse read
STS:
| GGAGCATGCC TATTCAGTAC AAGGACTCA ATTATGGAATC TACTTTGAAT TGTCG GCAGA ATGGTAGTCT CTTTGATGTC TCCCATATGT GTGGGCTGAG CCTCAAAGGG AAGGATTGCG TCCCTTTCCT TGAGAAGCTT GTGATTGCTG ATGTTGCTGG ACTT GCCCCA GGAACTGGGA CTCTTACAGT CTTTACAAAT GAAAATGGGG GTGCAATNG A TGATTCGGTT ATTACCAAGG TGAAGGATGA TCACATATAC CTTGTTGTGA ATG CTGGCTG CAGGGATAAG GATCTTGCTC ACATTGAGGC ACATATGAAG AGTTTCAC AG |
STS Forward / Reverse:
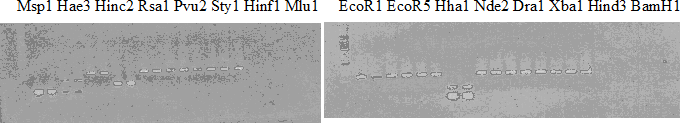
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): A255(Okitsu46gou)/G434(Kankitsu Chuukanbohon Nou5)Comment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-wase
Comment:
Figure legend:
 |
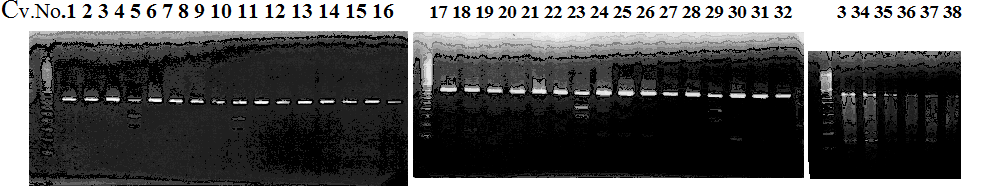
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: EcoR1Rank as CAPS marker: no homozygous cultivar with minor allele
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Experiments: CAPS pattern of Citrus collection
Experiments: Segregation pattern
Population name: Kiyomi x Miyagawa-waseRestriction enzyme: EcoR1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: EcoR1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: EcoR1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
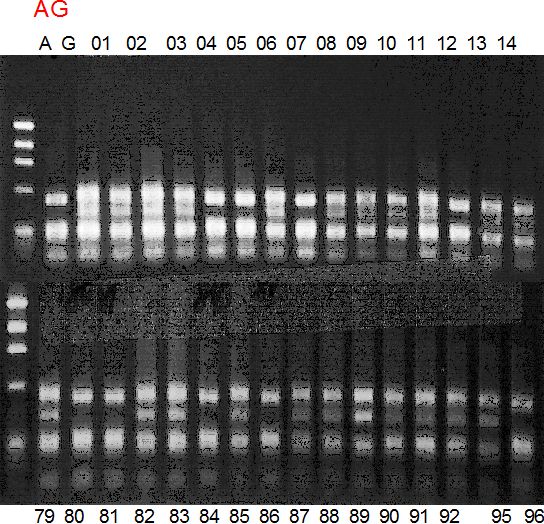
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Nde2
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10015433mScaffold: scaffold_2
Start: 35470078
End: 35472862
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00048:mRNA_13.1Scaffold: C_unshiu_00048
Start: 64652
End: 67377
Gene function: Aminomethyltransferase
AGI map info (Shimada et al. 2014)
AGI LG: AGI-06AGI cM: 18.136
KmMg map info (Omura et al. 2003)
KmMg LG:KmMg cM:
JtTr map info (Omura et al. 2003)
JtTr LG: JtR-06SJtTr cM: 67.371
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI130DNA sequence around SNP1:
| TATGTGTGGGCTGAGCCTCAAAGGGAAGGATTGCGTCCCTTTCCTTGAGAAGCTTGTGAT [T/C]GCTGATGTTGCTGGACTTGCCCCAGGAACTGGGACTCTTACAGTCTTTACAAATG AAAAT |
DNA sequence around SNP2:
DNA sequence around SNP3:
DNA sequence around SNP4: