Locus name: If0208
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 58Main product (bp): 600
Comment on PCR:
F-primer: AATATTTCTGCGAATCACTGA
R-primer: GCAAACCACACCAAGGA
EST info
EST Accession number 1: DC893504EST Accession number 2: DC893505
sequence-tagged site (STS seq)
STS:| TATCACAGGGCTCAGGGGATACTTGACTTCTGCTTTCTTCACGACGGGCTTAAAGGTTGT TGCCCCTCTCTTTGCCGTCTATGTAACTTGGCCAGTGCTTCGACTGCCAGCCTTGGTTGC AGTGCTTCCATTTCTGGTTGGTTGTGCGGCTCAACTTGCATTTGAGACCAATCTTGACAA GCGTGGGTCATCATGTTGGCCCTTAATTCCTATCATCTTTGAGGTTGTTTCTTGACTCAT ATTTATCTTTCTCAGGCCAAAATCAAGGATTTATTAACTTTATTTTAAAATCAAGGATTT TAATGACTTTATTTTATTGGCAGGTATACAGACTTTATCAGTTGAGCAAAGCTGCTAATT TCATTGAGAGATTGATGTTCTCAATGAAAGATCTTCCGAGATCTCCGGAATTGCTGGAAA GAGGCAGTGCAATGGTTTCCATGGTAGTGATTTTCCAA |
STS Forward / Reverse: Foward read
STS:
| GGTGGGGTCTTATCACAGGGCTCAGGGGATACTTGACTTCTGCTTTCTTCACGACGGGCT TAAAGGTTGTTGCCCCTCTCTTTGCCGTCTATGTAACTTGGCCNGTGCTTCGACTGCCAG CCTTGGTTGCAGTGCTTCCATTTCTGGTTGGTTGTGCGGCTCAACTTGCATTTGAGACCA ATCTTGACAAGCGTGGGTCNTCATGTTGGCCCTTAATTCCTATCATCTTTGAGGTTGTTT CTTGACTCATATTTNTCTTTCTCAGGCCAAAATCAAGGATTTNTTAACTTTATTTTAAAA TCAAGGATTTTAATGACTTTATTTTATTGGCAGGTATACAGACTTTATCAGTTGAGCAAA GCTGCTAATTTCATTGAGAGATTGATGTTCTCAATGAAAGATCTTCCGAGATCTCCGGAA TTGCTGGAAAGAGGCAGTGCAATGGTTTCCATGGTAGTGATTTTCCAA |
STS Forward / Reverse: Foward read
STS:
| CTCGGAAGATCTTTCATTGAGAACATCAATCTCTCAATGAAATTAGCAGCTTTGCTCAAC TGATAAAGTCTGTATACCTGCCAATAAAATAAAGTCATTAAAATCCTTGATTTTAAAATA AAGTTAATAAATCCTTGATTTTGGCCTGAGAAAGAAAAATATGAGTCAAGAAACAACCTC AAAGATGATAGGAATTAAGGGCCAACATGATGACCCACGCTTGTCAAGATTGGTCTCAAA TGCAAGTTGAGCCGCACAACCAACCAGAAATGGAAGCACTGCAACCAAGGCTGGCAGTCG AAGCACTGGCCAAGTTACATAGACGGCAAAGAGAGGGGCAACAACCTTTAAGCCCGTCGT GAAGAAAGCAGAAGTCAAGTATCCCCTGAGCCCTGTGATAAGACCCCACCTCTTTGGGCT GAATTGTAAA |
STS Forward / Reverse: Reverse read
STS:
| AATATTTCTGCCAATCACTGATCGAGTTCAGAGGCCATATTTACAATTCAGCCCAAAGAG GTGGGGTCTTATCACAGGGCTCAGGGGATACTTGACTTCTGCTTTCTTCACGACGGGCTT AAAGGTTGTTGCCCCTCTCTTTGCCGTCTATGTAACTTGGCCAGTGCTTCGACTGCCAGC CTTGGTTGCAGTGCTTCCATTTCTGGTTGGTTGTGCGGCTCAACTTGCATTTGAGACCAA TCTTGACAAGCGTGGGTCNTCATGTTGGCCCTTAATTCCTATCATCTTTGAGGTTGTTTC TTGACTCATATTTNTCTTTCTCAGGCCAAAATCAAGGATTTATTAACTTTATTTTAAAAT CAAGGATTTTAATGACTTTATTTTATTGGCAGGTATACAGACTTTATCAGTTGAGCAAAG CTGCTAATTTCATTGAGAGATTGATGTTCTCAATGAAAGATCTTCCGAGATCTCCGGAAT TGCTGGAAAGAGGCAGTGC |
STS Forward / Reverse: Reverse read
STS:
| GGTGGGGTCT TATCACAGGG CTCAGGGGAT ACTTGACTTC TGCTTTCTTC ACGAC GGGCT TAAAGGTTGT TGCCCCTCTC TTTGCCGTCT ATGTAACTTG GCCTGTGCTT CGACTGCCAG CCTTGGTTGC AGTGCTTCCA TTTCTGGTTG GTTGTGCGGC TCAA CTTGCA TTTGAGACCA ATCTTGACAA GCGTGGGTCATCATGTTGGC CCTTAATTC C TATC |
STS Forward / Reverse:
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-waseComment:
Figure legend:
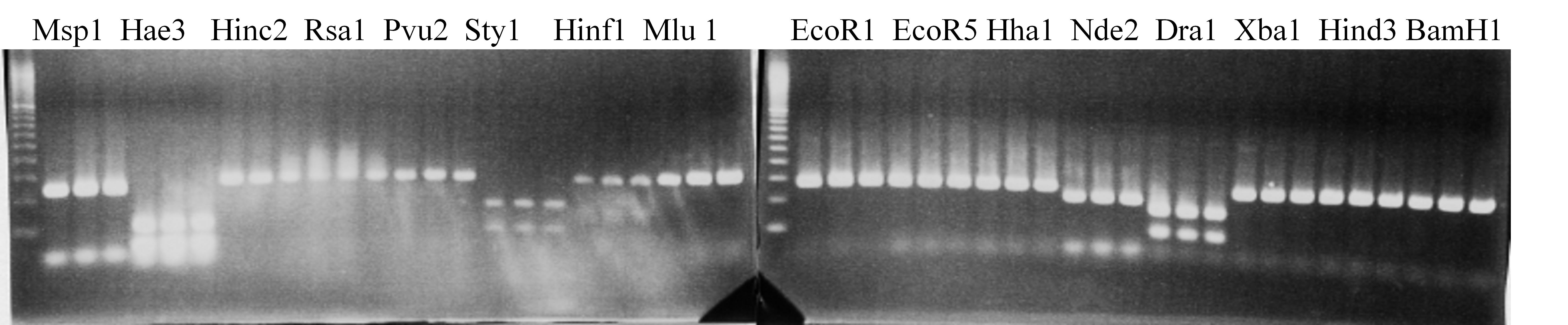 |
Compared cultivars (Left, (Middle), Right): A255(Okitsu46gou)/G434(Kankitsu Chuukanbohon Nou5)
Comment:
Figure legend:
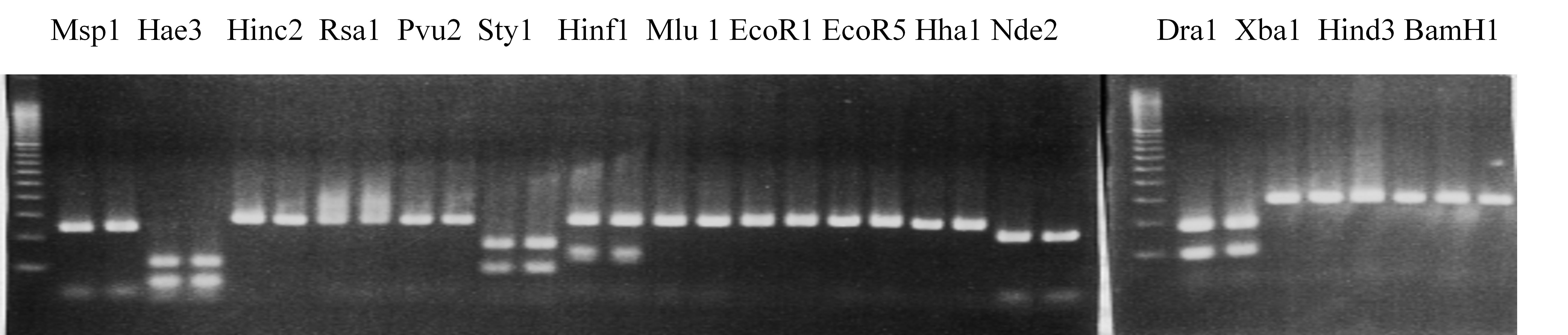 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Hinf1Rank as CAPS marker: Available marker
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
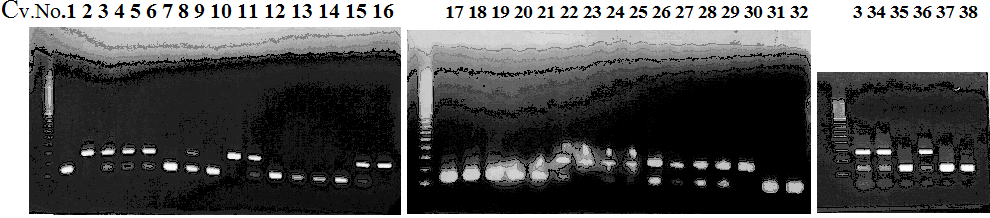 |
Experiments: CAPS pattern of Citrus collection
Experiments: Segregation pattern
Population name: Kiyomi x Miyagawa-waseRestriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
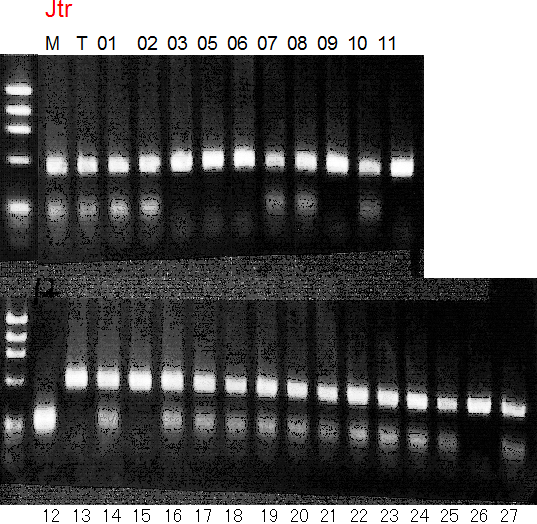 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
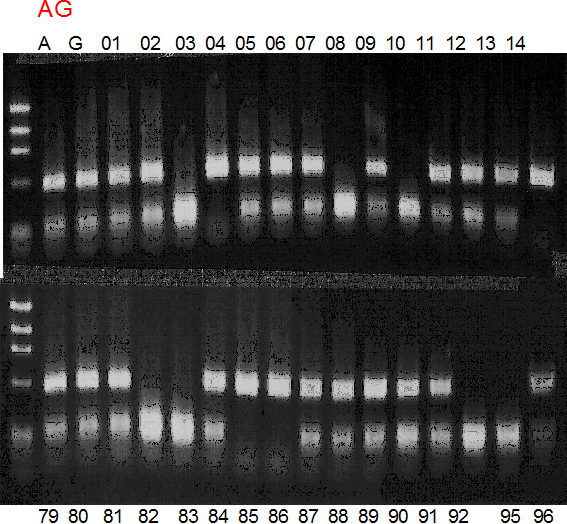 |
Population name: Kiyomi x Okitsu41gou
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
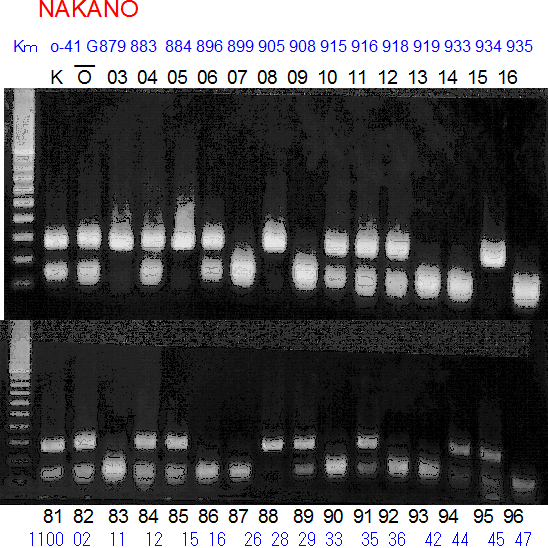 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10001680mScaffold: scaffold_5
Start: 40628244
End: 40630241
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00234:mRNA_9.1Scaffold: C_unshiu_00234
Start: 94988
End: 99176
Gene function: TRNA-processing ribonuclease BN
AGI map info (Shimada et al. 2014)
AGI LG: AGI-09AGI cM: 39.296
KmMg map info (Omura et al. 2003)
KmMg LG: KmG-09KmMg cM: 72.036
JtTr map info (Omura et al. 2003)
JtTr LG: JtR-09SJtTr cM: 115.15
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG: NAT-09Km/O41 cM: 89.867
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K: M,KSNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI083DNA sequence around SNP1:
| AGTCAAGAAACAACCTCAAAGATGATAGGAATTAAGGGCCAACATGA[T/C]GACCCACG CTTGTCAAGATTGGTCTCAAATGCAAGTTGAGCCGCACAACCAACCAGAAAT |
DNA sequence around SNP2:
| TTTCTTCACGACGGGCTTAAAGGTTGTTGCCCCTCTCTTTGCCGTCTATGTAACTTGGCC [A/T]GTGCTTCGACTGCCAGCCTTGGTTGCAGTGCTTCCATTTCTGGTTGGTTGTGCGG CTCAA |
DNA sequence around SNP3:
DNA sequence around SNP4: