Locus name: Lp0143
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 58Main product (bp): 1800
Comment on PCR:
F-primer: ATTATGAGTGACCCGTTTGTT
R-primer: TGTTGTTCGGCGATGA
EST info
EST Accession number 1: AU300576EST Accession number 2: DC893258
sequence-tagged site (STS seq)
STS:| GTATGCTGGGTCGCTTTCTGCAGGATCTTATGTGCTGAACGCAAACAAAGGAAAGAAGGA GAGAATTGGTAGACTTTTGGAAATGCATGCCAACAGTAGGGAGGATGTAAAGGTTGCCTT AGCAGGTGACATCATTGCTCTTGCAGGTCTTAAAGACACCATTACTGGTGAAACACTTTG TGATGCAGACCATCCAATTCTGCTTGAACGAATGGACTTCCCCGATCCTGTGATTAAGGT TGCAATTGAGCCCAAGACTAAAGCTGATATTGACAAAATGGCAAATGGCCTGATCAAACT TGCTCAAGAAGACCCTTCATTCCACTTCTCACGAGANGAAGAGATCAACCAGACAGTGAT TGAGGGTATGGGAGAATTGCATCTTGAGATTATCGTCGATCGCCTAAAGAGGGAATTTAA GGTATATATTGGCCCCTCCATACATCTGAGTATCATGTTAATGTTAGGTATTTTTTTACT TCACTTTATTGGGTTTAAACCTTATTCATGGTACTTTTTTCTTGGAATATATGATTGATN TTAGTGTATAGGTATATACTTTTACATACAATACTTGTACGCATATCAGTTGTTGTGAAT ATCAGTATGCACAGTGATATAGTTTTATGTCNGTGAGCAGAAGAGAACTAAAAGATGCAA TAAAATTGTTTGACTGATGCTATTCTTATTTCCTGCAAATTGCAAAGAAAAATATGTTCT GGATGCATAGACTTTTTCTTTGTCTTAACTGATGATCTTTGGGGCTAATA |
STS Forward / Reverse: Foward read
STS:
| TGTGCTGAACGCAAACAAAGGAAAGAAGGAGAGAATTGGTAGACTTTTGGAAATGCATGC CAACAGTAGGGAGGATGTAAAGGTTGCCTTAGCAGGTGACATCATTGCTCTTGCAGGTCT TAAAGACACCATTACTGGTGAAACACTTTGTGATGCAGACCATCCAATTCTGCTTGAACG AATGGACTTCCCCGATCCTGTGATTAAGGTTGCAATTGAGCCCAAGACTAAAGCTGATAT TGACAAAATGGCAAATGGCCTGATCAAACTTGCTCAAGAAGACCCTTCATTCCACTTCTC ACGAGATGAAGAGATCAACCAGACAGTGATTGAGGGTATGGGAGAATTGCATCTTGAGAT TATCGTCGATCGCCTAAAGAGGGAATTTAAGGTATATATTGGCCCCTCCATACATCTGAG TATCATGTTAATGTTAGGTATTTTTTTACTTCACTTTATTGGGTTTAAACCTTATTCATG GTACTTTTTT |
STS Forward / Reverse: Foward read
STS:
| GATTTGAGCCTATGGAGGCAGGAAAGGGGATATGAGTTCAAGAGTGAGATCAAGGGAGGT GCAGTGCCAAAAGAATATATTCCTGGGGTGATGAAAGGACTAGAAGAATGCATGAGCAAT GGTGTGCTTGCAGGCTTCCCTGTTGTTGACGTACGTGCTGCACTAGTTGATGGTTCCTAC CATGATGTTGATTCTAGTGTCTTGGCATTTCAACTTGCGGCCAGGGGAGCTTTCAGGGAG GGCATGAGAAAGGCTGGCCCTAAAATGCTGGAACCTATAATGAAAGTCGAAGTTGTCACA CCTGAAGAACACTTGGGAGATGTGATTGGTGATCTCAACTCGAGGAGAGGTCAGATCANC AGTTTTGGTGACAAACCTGGTGGTTTGAAGGTACATTGAATTTCTTTCTCAGCACTCTGT CCGGATATACATCCATCTGGTTAATTCCTAAGTGTCTTTGATTCTTGATCTTTCTTTCCT TAGTTTTTCCCGCTGCAGTCTAAATTTTNGATTCATGGGATATTTTGGTATTATCGCAGG TGGTTGATGCGCTGGTTGCCACTGGCAGAAATGTTNCAATATGTTAGTGCACTCCCGAGG GATNACAAAAGGCCGTGCA |
STS Forward / Reverse: Reverse read
STS:
| GAGAATTGGT AGACTTTTGG AAATGCATGC CAACAGTAGG GAGGATGTAA AGGTT GCCTT AGCAGGTGAC ATCATTGCTC TTGCAGGTCT TAAAGACACC ATTACTGGTG AAACACTTTG TGATGCAGAC CATCCAATTC TGCTTGAACG AATGGACTTC CCCG ATCCTG TGATTAAGGT TGCAATTGAG CCCAAGACTA AAGCTGATAT TGACAAAAT G GCAAATGGC |
STS Forward / Reverse:
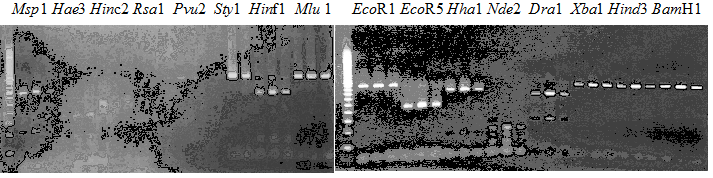
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-waseComment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): A255(Okitsu46gou)/G434(Kankitsu Chuukanbohon Nou5)
Comment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Hinc2Rank as CAPS marker: Others
Comment:
Figure legend:
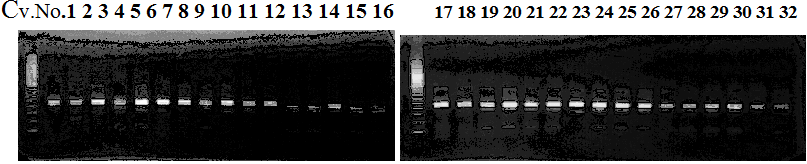
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Resriction enzyme: Nde2
Rank as CAPS marker: no homozygous cultivar with minor allele
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Experiments: CAPS pattern of Citrus collection
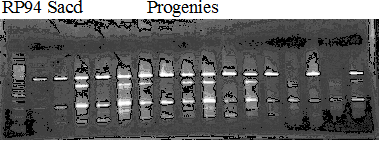
Experiments: Segregation pattern
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)Restriction enzyme: Hinc2
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Mbo1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese Acidless
Restriction enzyme: NC
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10019139mScaffold: scaffold_3
Start: 6076949
End: 6080875
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00159:mRNA_40.1Scaffold: C_unshiu_00159
Start: 339926
End: 343475
Gene function: Translation elongation factor G
AGI map info (Shimada et al. 2014)
AGI LG: AGI-05AGI cM: 45.297
KmMg map info (Omura et al. 2003)
KmMg LG:KmMg cM:
JtTr map info (Omura et al. 2003)
JtTr LG: JtR-05JtTr cM: 18.056
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1:DNA sequence around SNP1:
DNA sequence around SNP2:
DNA sequence around SNP3:
DNA sequence around SNP4: