Locus name: Mf0010
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 56Main product (bp): 400
Comment on PCR:
F-primer: AATGTTATCATCAATGGCAC
R-primer: ATACCAGCCTTGGCTTCTCC
EST info
EST Accession number 1: C81689EST Accession number 2:
sequence-tagged site (STS seq)
STS:| TGTCAAACCCACCAAAACCACCATTCTTCACCAAAATAGAACCAAACCCATTTCAAGAAA CCAAAGGCTCGTTGCTTTAGCAACCCCAGAATCACTCACAGCTGAAGAAGCAACAACAAC ATCCACAGAGACCATCCCAGCAACCCCAGTAGAAACTAAGGTCCAAACCAAATCTCTCTC TCCCTCTCTACATATGTGTGTTTATGTGTACTGTTTC |
STS Forward / Reverse: Foward read
STS:
| GCTCCTCATTCCCTTCCCAAAAACTCACTGTCAAACCCACCAAAACCACCATTCTTCACC AAAATAGAACCAAACCCATTTCAAGAAACCAAAGGCTCGTTGCTTTAGCAACCCCAGAAT CACTCACAGCTGAAGAAGCAACAACAACATCCACAGAGACCATCCCAGCAACCCCAGTAG AAACTAAGGTCCAAACCAAATCTCTCTCTCCCTCTCTACATATGTGTGTTTATGTGTACT GTTTCTATGTGTACTGTTTCACTTAATAAAAGTTGTGTGTTTTCTTTTTCTTTTCTTCTG GGCAGAAGGCGGGGGTGTCAGTGAAGCAATTGGAGAAGCCAGGGCTGGTAT |
STS Forward / Reverse: Foward read
STS:
| TTTGGTTTGGACCTTAGTTTCTACTGGGGTTGCTGGGATGGTCTCTGTGGATGTTGTTGT TGCTTCTTCAGCTGTGAGTGAAACTGGGGTTGCTAAAGCAACGAGCCTTTGGTTTCTTGA AATGGGTTTGGTTCTATTTTGGTGAAGAATGGTGGTTTTGGTGGGTTTGACAGTGAGTTT TTGGGAAGGGAATGAGGAGCGTGTGGTGAAT |
STS Forward / Reverse: Reverse read
STS:
| AAACCACCAT TCTTCACCAA AATAGAACCA AACCCATTTC AAGAAACCAA AGGCT CGTTG CTTTAGCAAC CCCAGAATCA CTCACAGCTG AAGAAGCAAC AACAACATCC ACAGAGACCA TCCCAACAAC CCCAGTAGAA ACTAAGGTCC AAACCAAATC TCTC TCTCCC TCTCTACATA TGTGTGTTTA TGTGTACTGT TTC |
STS Forward / Reverse:
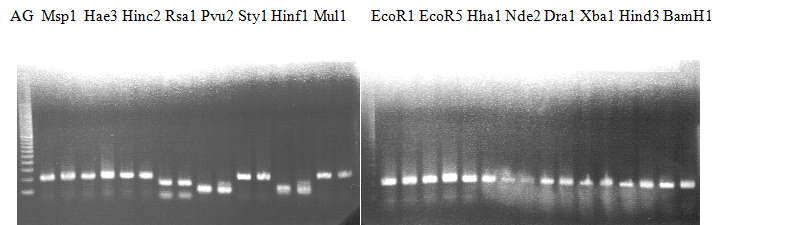
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): A255(Okitsu46gou)/G434(Kankitsu Chuukanbohon Nou5)Comment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Miyagawa-wase/Trovita orange
Comment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Hinf1Rank as CAPS marker: Others
Comment:
Figure legend:
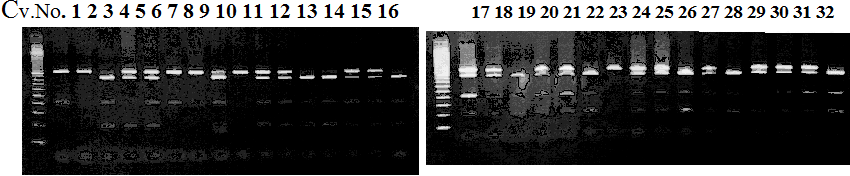
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
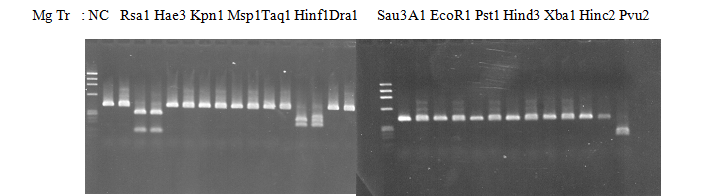
Resriction enzyme: Pvu2
Rank as CAPS marker: no homozygous cultivar with minor allele
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Resriction enzyme: NC
Rank as CAPS marker: Others
Comment: PCR_condition: 52C aneal
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Experiments: CAPS pattern of Citrus collection
Experiments: Segregation pattern
Population name: Jutaro unshu x Trovita orangeRestriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Okitsu41gou
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Pvu2
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Pvu2
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10032902mScaffold: scaffold_4
Start: 19467669
End: 19468803
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00099:mRNA_4.1Scaffold: C_unshiu_00099
Start: 12446
End: 13357
Gene function: 30S ribosomal protein 3, chloroplastic
AGI map info (Shimada et al. 2014)
AGI LG: AGI-07AGI cM: 55.503
KmMg map info (Omura et al. 2003)
KmMg LG: KmG-07KmMg cM: 42.271
JtTr map info (Omura et al. 2003)
JtTr LG: JtR-07JtTr cM: 35.215
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG: NAT-07Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI048DNA sequence around SNP1:
| AAGCAACAACAACATCCACAGAGACCATCCCAACAACCCCAGTAGAAACTAAGGTCCAAA [T/C]CAAATCTCTCTCTCCCTCTCTACATATGTGTGTTTATGTGTACTGTTTC |
DNA sequence around SNP2:
| GCAACCCCAGAATCACTCACAGCTGAAGAAGCAACAACAACATCCACAGAGACCATCCCA [A/G]CAACCCCAGTAGAAACTAAGGTCCAAA |
DNA sequence around SNP3:
DNA sequence around SNP4: