Locus name: Mf0039
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 56Main product (bp): 1500
Comment on PCR:
F-primer: CCAATTCCTCAATATATGCAG
R-primer: CACAAGTATACTGCTGTACCA
EST info
EST Accession number 1: C81786EST Accession number 2: DC893617
sequence-tagged site (STS seq)
STS:| CAACTCTCTGCCTTTTCATGTGCAAGCTAGCCCTGTCAGTCTTTTGTCTCTGGACTATAG CAGCAAGTCTACCCACATTTTTATGTCACAAAGGAATGCGCTTGGGTATGTTAGAGACAG TTATCTACAGGAAGCAGAAATGGAAATGAATTCATTTGTCATGATCCTTTTGATATTAGT CTACACAGTTGATCTTTCTAGTTAATTTTTACCAAAAATCACATGTGATTTAGCTCTGCN GATGTCTAGGTTTATGGCCACTAGATATGCATTACCCCTTNTTTTGGCTTCTAAGGATAA AGGTGTTTTTATTGGTTCCTTTGTTGCTTAACCNTCTCATCTTGTACCTCTTGGATAACT TATGAAGTATACAGCATGCTTTCTTGTGAGAAAATATTATAATCCCCATTTACTGGGTTT TCAGTTTATAAACCAATGTAGCAGTTGTAAATAGCTTCTGTTTTGTTAATTATTGAAACC ATTTCTTTTTNAACATCCAGGTTTTTTGCTGCTTTGCCAATTCGGTGGCCTCTACGCGCT TTCTTTTGCAAGATGAATTTAACATCCAGACAACTGCCTGTGATAATTGCATCATTGTAC GGTCACTTACATTATCTATTAACACTATTGCTATAGTTACACATCTTTGNTTATTTGTTA TATGNGTTTCTTATTTGTATGCAGGGGTTTATGTTTTGCCTCAGCC |
STS Forward / Reverse: Foward read
STS:
| GTCAGTCTTTTGTCTCTGGACTATAGCAGCAAGTCTACCCACATTTTTATGTCACAAAGG AATGCGCTTGGGTATGTTAGAGACAGTTATCTACAGGAAGCAGAAATGGAAATGAATTCA TTTGTCATGATCCTTTTGATATTAGTCTACACAGTTGATCTTTCTAGTTAATTTTTACCA AAAATCACATGTGATTTAGCTCTGCCGATGTCTAGGTTTATGGCCACTAGATATGCATTA CCCCTTCTTTTGGCTTCTAAGGATAAAGGTGTTTTTATTGGTTCCTTTGTTGCTTAACCC TCTCATCTTGTACCTCTTGGATAACTTATGAAGTATACAGCATGCTTTCTTGTGAGAAAA TATTATAATCCCCATTTACTGGGTTTTCAGTTTATAAACCAATGTAGCAGTTGTAAATAG CTTCTGTTTTGTTAATTATT |
STS Forward / Reverse: Foward read
STS:
| TTCTTTGCAAGATGAATTAACATCCAGACAACTGCCTGTGATAATTGCATCATTGTACGT CACTTACATTATCTATTANCACTATTGCTATAGTTACACATCTTGGTTATTNGTTATATG TGTTTCTTATTGGTATGCAGGGGTTTATGTTTTGCCTCAGCCAACTTGCATGCATATTTT CCTTAGTTGCATGCATCGTGGGAAGTGATGAACTTCAGGAGGCTTCACAGATACTGAATC TATTGGCTGATTTAGTTTATTGCACGTAAGTGTTTTCCTTCTTTCCACACTGAAAGCTCC TAGGCAAAATGGCTGCAAAATATCCTACTGATTCTGCTTTATGTTTTGGTTTGCAGGGTC TGTGCCTGCATGCAGGTATGCAATTNATTCAACCACATGCAAATGAAATTACATTATAAC GAAATATTTCTTCACAGTTTATTTGAATTGTCTGCAGACACAGCACAAGGTTGAAATGGA CAAAAGGGATGGTAAATTTGGTCCACAACCAATGGCGGTACCCCCTCATCAACAGATGTC TCGCATTGATCAGCCATATCCTGCGACGGTTGGATATCCACCTCAACAACAAGCATATGG TTATCCTCCTCAAGGCTATCCACCACCTCAGGGTCAGGGCTACCCACCACCCCAAGGTCA AGGCTATCCAGCTGGAGGTTATCCCCCGCCTGCATACAATCAAGGCCATTACCCACCTCC TACCCATTAAGGCCTAGTTCCGCTGTGGTGCATGTTTGATATTGGTTTGTGAAAATGAAC TTTTGAGAC |
STS Forward / Reverse: Reverse read
STS:
| TCAAACATGCACCACAGCGGAACTAGGCCTTAATGGGTAGGAGGTGGGTAATGGCCTTGA TTGTATGCAGGCGGGGGATAACCTCCAGCTGGATAGCCTTGACCTTGGGGTGGTGGGTAG CCCTGACCCTGAGGTGGTGGATAGCCTTGAGGAGGATAACCATATGCTTGTTGTTGAGGT GGATATCCAACCGTCGCAGGATATGGCTGATCAATGCGAGACATCTGTTGATGAGGGGGT ACCGCCATTGGTTGTGGACCAAATTTACCATCCCTTTTGTCCATTTCAACCTTGTGCTGT GTCTGCAGACAATTCAAATAAACTGTGAAGAAATATTTCGTTATAATGTAATTTCATTTG CATGTGGTTGAATTAATTGCATACCTGCATGCAGGCACAGACCCTGCAAACCAAAACATA AAGCAGAATCAGTAGGATATTTTGCAGCCATTTTGCCTAGGAGCTTTCAGTGTGGAAAGA |
STS Forward / Reverse: Reverse read
STS:
| GTTATCTACA GGAAGCAGAA ATGGAAATGA ATTCATTTGT CATGATCCTT TTGAT ATTAG TCTACACAGT TGATCTTTCT AGTTAATTTT TACCAAAAAT CACATGTGAT TTAGCTCTGC TGATGTCTAG GTTTATGGCC ACTAGATATG CATTACCCCT TATT TTGGCT TCTAAGGATA AGGTGTTTT TATTGGTTCC TTTGTTGCTT AACCCTCTCA TCTTGT |
STS Forward / Reverse:
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-waseComment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Miyagawa-wase/Trovita orange
Comment: PCR_condition: 48C aneal
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Hinf1Rank as CAPS marker: Available marker
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Experiments: CAPS pattern of Citrus collection
Rsetriction enzyme: Hha1Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
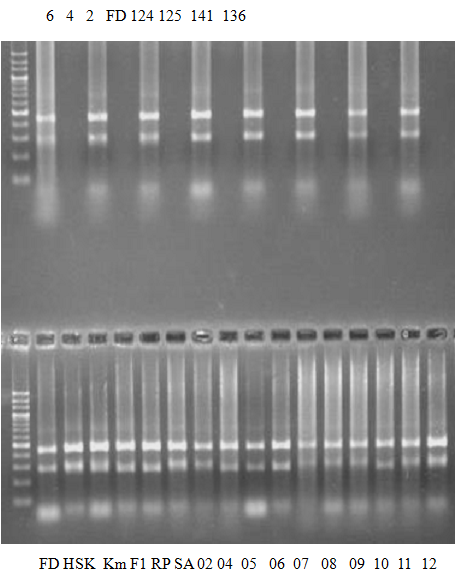
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
 |
Rsetriction enzyme: Hinf1
Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
 |
Experiments: Segregation pattern
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese AcidlessRestriction enzyme: Hha1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Okitsu41gou
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID:Scaffold:
Start:
End:
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00343:mRNA_25.1Scaffold: C_unshiu_00343
Start: 221381
End: 224081
Gene function: PLAC8 family protein
AGI map info (Shimada et al. 2014)
AGI LG: AGI-05AGI cM: 5.51
KmMg map info (Omura et al. 2003)
KmMg LG: KmG-05SKmMg cM: 5.643
JtTr map info (Omura et al. 2003)
JtTr LG: JtR-05JtTr cM: 10.014
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI016DNA sequence around SNP1:
| TTTTGGCTTCTAAGGATAAAGGTGTTTTTATTGGTTCCTTTGTTGCTTAACC[A/C]TCT CATCTTGTACCTCTTGGATAACTTATGAAGTATACAGCATGCTTTCTTGTGAGAAAA |
DNA sequence around SNP2:
| TCAAATAAACTGTGAAGAAATATTTCGTTATAATGTAATTTCATTTGCATGTGGTTGAAT [T/G]AATTGCATACCTGCATGCAGGCACAGACCCTGCAAACCAAAACATAAAGCAGAAT CAGTA |
DNA sequence around SNP3:
DNA sequence around SNP4: