Locus name: Mf0079
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 60Main product (bp): 1100
Comment on PCR:
F-primer: GGTGACTTTGCGGCCAAGGTC
R-primer: GGCAGTGGCTCATCTCAACGA
EST info
EST Accession number 1: C81861EST Accession number 2: DC893647
sequence-tagged site (STS seq)
STS:| GGCCTGATTGACATTCCAAGGTACGAGNTTGACTCACTCCTATACCTACATCATAAATAA TTTTAAAAAAAAAATTGCGTGCACAAGTAGGTTTGCAACAGAAATAATGTTCATGTTTTT TTATAATCCATGAAAAAATATTTGCAACAGAAACAATGTTCATCTCTTTTTATGATCCAT GAAAACATATCAGAAATTTGTCTCAAATTTATATAATCATATTGCATGCCAAAGACAAAT GATCTAGACATGGTAATGCTGAAACGGGAAGATATAAAAGCTTCATGACTCACTTCTGTT TAGCAATTTATGAACAGCACTTCTGCTTGTTGATCAATGTCAAATATCTGAACTGCTATG CAAGAAGGCTATTTTTGCCGTACTTGATCCTATGATTTGCCAAAGTCTAGCTTTCTTATG TAAAATATGACAGTGCCTCAGCTGTTTTATCACGTGGGTAACCAGTACNCATACATACAC ACTGCCTTCNCACTGAGTTCTTTTCTTCCTAATCTCTTAAGCCTCTAAATCTTTCTTGGT TTAAGTTCTCTTTTTACAATTGTATTGCTAGAGATCATTAAAAGTTTTAGTCATTATGTC TGGTTAATTATCGACGTTGAAATTTTCTACATGGATTTCAATCACCCTTCTAGATAATTT GATGTGTATGGTGAATTGCAAGTTCTTGATAAATTTATAATGCTTCTAAGCTGTATTTTC CTGCATTTTCATGTAGACTGCCAAAGACACCCACCTGGGTTATGAGGACCAGGATGGAGC AAAAGCTAT |
STS Forward / Reverse: Foward read
STS:
| TGTCATCTCTTTTATGATCCATGAAAACATATCAGAAATTGTCTCAAAATTATATAATCA TATTGCATGCCAAAGACAAATGATCTAGACATGGTAATGCTGAAACGGAAGATATAAAAG CTTCATGACTCACTTCTGTTTAGCAATTTATGAACAGCACTTCTGCTTGTTGATCAATGT CAAATATCTGAACTGCTATGCAAGAAGGCTATTTTTGCCGTACTTGATCCTATGATTTGC CAAAGTCTAGCTTTCTTATGTAAAATATGACAGTGCCTCAGCTGTTTTATCACGTGGGTA ACCAGTACGCATANATACACACTGCCTTCNCACTGAGTTCTTTTCTTCCTAATCTCTTAA GCCTCTAAATCTTTCTTGGTTTAAGTTCTCTTTTTACAATTGTATTGCTAGAGATCATTA AAAGTTTTAGTCATTATGTCTGGTTAATTATCGACGTTGAAATTTTCTACATGGATTTCA ATCACCCTTCTAGATAATTTGATGTGTATGGTGAATTGCAAGTTCTTGATAAATTTATAA TGCTTCTAAGCTGTATTTTCCTGCATTTTCATGTAGACTGCCAAAAGACACCCAACCTGG GTTATTGAGGACCAAGGATGGAGCCAAAAAGCTAATGGACTTGGGTTTACAATTCATTCC CATGGATCAAATCATCAAGGACTCTNTCGAGAGTTTGAAGGCCAAGGGATTTATCTCTTG ANTGAAAGCTGCAACGTGTCAGCAGCTGCTGATGAACCCATCCGCTTTATNCAAGCAGAG GGCTGAGAGCTATAGTATAATTAGATAATT |
STS Forward / Reverse: Reverse read
STS:
| AATTATACTATAGCTCTCAGCCCTCTGCTTGA/ATAAAGCGGATGGGTTCATCAGCAGCT GCTGACACGTTGCAGCTTTCATTCAAGAGATAAATCCCTTGGCCTTCAAACTCTCGATAG AGTCCTTGATGATTTGATCCATGGGAATGAATTGTAAACCCAAGTCCATTAGCTTTTTGG CTCCATCCTTGGTCCTCAATAACCCAGGTTGGGTGTCTTTTGGCAGTCTACATGAAAATG CAGGAAAATACAGCTTAGAAGCATTATAAATTTATCAAGAACTTGCAATTCACCATACAC ATCAAATTATCTAGAAGGGTGATTGAAATCCATGTAGAAAATTTCAACGTCGATAATTAA CCAGACATAATGACTAAAACTTTTAATGATCTCTAGCAATACAATTGTAAAAAGAGAACT TAAACCAAGAAAGATTTAGAGGCTTAAGAGATTAGGAAGAAAAGAACTCAG |
STS Forward / Reverse: Reverse read
STS:
| AAGTTCTCTTTTTCCAATTGTATTGCTAGAGATCATTAAAAGTTTTAGTCATTATGTCTG GTTAATTATCGACGTTGAAATTTTCTACATGGATTTCAATCACCCTTCTAGATAATGTGA TGTTTATGGTGAATTGCAAGTTCTTGATAAATTTATAATGCTTCTAAGCTGTATTTTCCT GCATTTACATGTAGACTGCCAAAAGACACCCAACCTGGGTTATTGAGGACCAAGGATGGA GCCAAAAAGCTAATGGACTTGGGTTTACAATTCATTCCCATGGATCAAATCATCAAGGAC TCTGTCGAGAGTTTGAAGGCCAAAGGATTTATCTCTTGAATGAAAGCTGCAACGTGTCAG CAGCTGCTGAT |
STS Forward / Reverse: Reverse read
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-waseComment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Hind3Rank as CAPS marker: Available marker
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Resriction enzyme: Pvu2
Rank as CAPS marker: Others
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
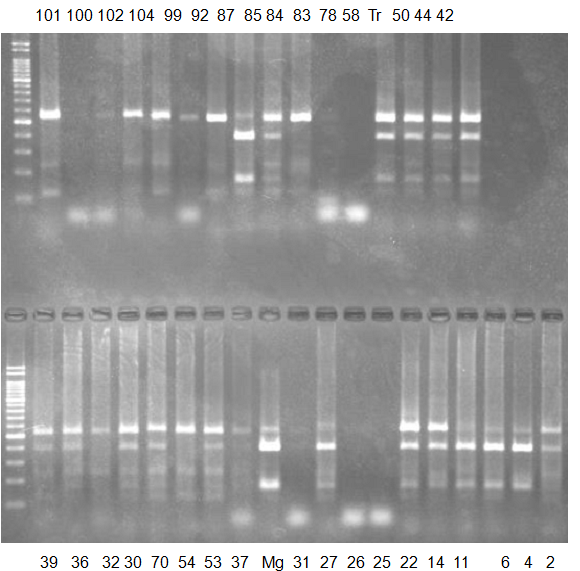
Experiments: CAPS pattern of Citrus collection
Rsetriction enzyme: Hind3Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
Rsetriction enzyme: NC
Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
Experiments: Segregation pattern
Population name: Kiyomi x Miyagawa-waseRestriction enzyme: Dra1
Comment: PCR_condition: 58C aneal
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Hind3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Hind3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Hind3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: NC
Comment: PCR_condition: 58C aneal
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
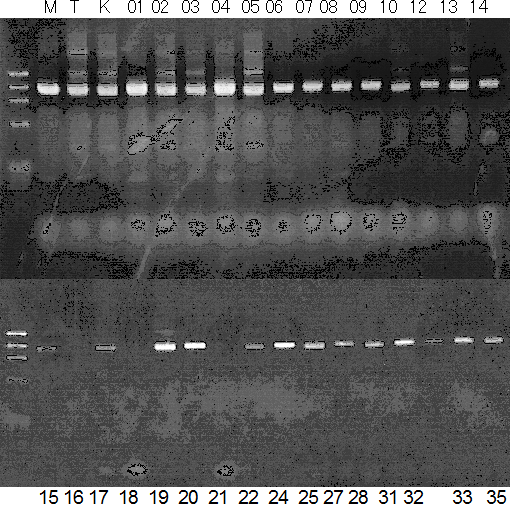 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Pvu2
Comment: PCR_condition: 58C aneal
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Okitsu41gou
Restriction enzyme: Pvu2
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Taq1
Comment: PCR_condition: 58C aneal
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
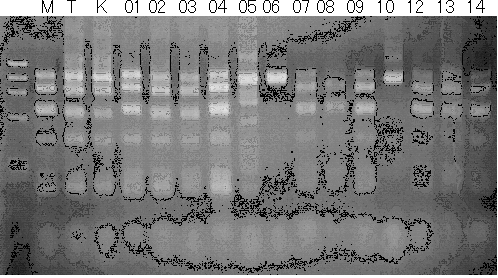 |
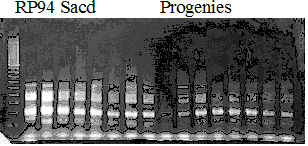
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese Acidless
Restriction enzyme: Xba1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10028839mScaffold: scaffold_8
Start: 24721937
End: 24724569
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00984:mRNA_8.1Scaffold: C_unshiu_00984
Start: 55676
End: 58212
Gene function: Cinnamoyl CoA reductase
AGI map info (Shimada et al. 2014)
AGI LG: AGI-08AGI cM: 3.409
KmMg map info (Omura et al. 2003)
KmMg LG: KmG-08KmMg cM:
JtTr map info (Omura et al. 2003)
JtTr LG:JtTr cM:
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG: NAT-082Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: Hb, CcCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI067DNA sequence around SNP1:
| TCAAGAGATAAATCCCTTGGCCTTCAAACTCTCGA[T/C]AGAGTCCTTGATGATTTGAT CCATGGGAATGAATTGTAAACCCAAGTCCATTAGCTTTTT |
DNA sequence around SNP2:
DNA sequence around SNP3:
DNA sequence around SNP4: