Locus name: Mf0092
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 58Main product (bp): 1800
Comment on PCR:
F-primer: GCTAAAGCCGGTTACACAGGC
R-primer: TGCTCTAGGAAATGGTTCCTC
EST info
EST Accession number 1: C81907EST Accession number 2: DC893668
sequence-tagged site (STS seq)
STS:| GGAAGAGGCCGAGTTGTCTGTTACATGATAACATTGGCAATGCAAGTATTGGNAGTTGGC ATGTGCCCCTTATTTGGCTTANCNGNCATGATTTGTTGTCAGATTTGCATAATCATTTGA AGTTGTAGCCCAATCATCGTCGGTTGGGTGTATGCATTACTCAAGCCANTCCTTTAGGAG AAATATTCATTAACATGTTGTCTGTTGCTATTACTATAATTCTGTTCTGATCAACTTTCT CCTTATTTGTCCATGTTTTGTGGTAAAATACTTTTTNTTTTTTTCCCCGGTTTCCTAAAN TTTTTGGGTTT |
STS Forward / Reverse: Foward read
STS:
| ACGTTGTAAATGAAAATCTGTTCCTTCTCTCTGTTCTGGTGGAAAGGTTAATCAAATTGG ATCTGTAACTGAGAGCATTGAAGCCGTCAGAATGTCCAAGCAAGCTGGGTGGGGTGTGAT GGCCAGCCACCGCAGTGGTGAGACTGAAGATACTTTCATTGCCGATCTCTCAGTGGGCTT GGCCACGGTAAATATGCTCTTTATATAAATGATACCATTGCTCAATTACAAGCAAAGTTT ATTATAAGACTTTTGTTTCTTATTGTTTTCTTGTTCATGATCAGGGTCAAATTAAGACAG GAGCACCATGCAGGTCAGAGCGNCTTGCAAAGTATAATCAGGTCAGCATCTTCTTATTTC ATCTTGAAAGATCATTTGACCTGTCTTCTTTTGCTTTTCCCAAATTTTAACGTTTCAGAA TTTGTGGGCTTGACTATAATTTAATATGTTCATGTAGCTTCTGAGAATTGAAGAAGAGCT CGGTGCTGAAGCAGTTTATGCTGGTGCCAAGTTCCGTGCCCCTGTTGAACCCTACTAGAG TTTGAGATGCTGATCTGGTGTGGTGCTGGATTTGAGCAATGGAGGTTTACTTCAATAAGT TATGTAAAATGGATTTTTATGGGTTTTAGC |
STS Forward / Reverse: Reverse read
STS:
| ATTTTACATAACTTATTGAAGTAAACCTCCATTGCTCAAATCCAGCACCACACCAGATCA GCATCTCAAACTCTAGTAGGGTTCAACAGGGGCACGGAACTTGGCACCAGCATAAACTGC TTCAGCACCGAGCTCTTCTTCAATTCTCAGAAGCTACATGAACATATTAAATTATAGTCA AGCCCACAAATTCTGAAACGTTAAAATTTGGGAAAAGCAAAAGAAGACAGGTCAAATGAT CTTTCAAGATGAAATAAGAAGATGCTGACCTGATTATACTTTGCAAGACGCTCTGACCTG CATGGTGCTCCTGTCTTAATTTGACCCTGATCATGAACAAGAAAACAATAAGAAACAAAA GTCTTATAAT |
STS Forward / Reverse: Reverse read
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-waseComment:
Figure legend:
 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Hha1Rank as CAPS marker: Available marker
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Experiments: CAPS pattern of Citrus collection
Experiments: Segregation pattern
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)Restriction enzyme: Hha1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Hha1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Hha1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
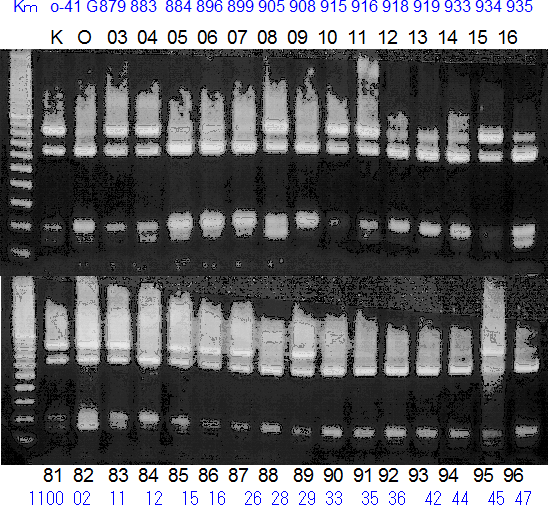
Population name: Kiyomi x Okitsu41gou
Restriction enzyme: Hha1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese Acidless
Restriction enzyme: NC
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10012056mScaffold: scaffold_6
Start: 21137783
End: 21142157
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00052:mRNA_27.2Scaffold: C_unshiu_00052
Start: 168316
End: 172622
Gene function: Phosphopyruvate hydratase
AGI map info (Shimada et al. 2014)
AGI LG: AGI-04AGI cM: 59.19
KmMg map info (Omura et al. 2003)
KmMg LG: KmG-04KmMg cM: 30.795
JtTr map info (Omura et al. 2003)
JtTr LG:JtTr cM:
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG: NAT-04Km/O41 cM: 11.576
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI138DNA sequence around SNP1:
| GGTCAAATGATCTTTCAAGATGAAATAAGAAGATGCTGACCTGATTATACTTTGCAAG[A /G]CGCTCTGACCTGCATGGTGCTCCTGTCTTAATTTGACCCTGATCATGAACAAGAAAA CAA |
DNA sequence around SNP2:
DNA sequence around SNP3:
DNA sequence around SNP4: