Locus name: Mf0096
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 60Main product (bp): 1200
Comment on PCR:
F-primer: GGAGTATCACCCCCACATGCT
R-primer: AAGTGCACAATGCTCCTTCTC
EST info
EST Accession number 1: C81915EST Accession number 2: DC893673
sequence-tagged site (STS seq)
STS:| CTCACTTCATGTATCCAAGGTCAGGGATCCAGTTGTTCTTTTATTGCTGATGGATTAATT TATTGCACAAACAGAAAGTGAGGAGATTTTATTTAGGTGCCATGCACGACTGTAAGGTGT TTAGGCCCCATCTTTGAGTATGAGAAAGTAATGGAACTACCAGCTACTGGTGGTCCCTTT AAAATGCCTTCNTTGATTTAAACATATAATGTACATAGGATGTTCCCCACAAGAAAAGTG CAGGCCATGAGAGCTGACTATTACTCACTGTAATGACATTATTATATTTATTTATTACTC AACTTTGAAAGTAATTCAATGATGTCTTCCCTTAAGAACTTTCATGTGTGTAAGTATGAC GTAATTGTCTATCAGCTAAATTGACTGTGTTGTTGAGAAATTACCCCATTGAATGTTCTA GGATCTCATTGACCTCTGACTCTGGGTTTTACATTGACAGTGGAGTTGACCAGTTTAAGC AGCAATTTGCCCGCCTTGAGGAGCAATATGGTAAAAGAGAAAGACATATTCCACTCCGAA GAAAGTATACCTCCTTGCCTAGGTATGGGTGATGCCAAAATTCTGAGCAGTTATTAGGTC ATTATCATTTTTGTTTAATTAGTTGCTGCCATTTTCTTCTCCACAGGGAAAGAATCAGTA CATCTGAAGATGAGGCTACAGATCAAACAGATTCTTTGAAGAAACGTACAGCTTCTGTCA CTCGTGCTACTCTCCAGAGCCCTCCAGTTTGCCCAGGTTTGCTGAAT |
STS Forward / Reverse: Foward read
STS:
| AGTTGTTCTTTTATTGCTGATGGATTAATTTATTGCACAAACAGAAAGTGAGGAGATTTT ATTTAGGTGCCATGCACGACTGTAAGGTGTTTAGGCCCCATCTTTGAGTATGAGAAAGTA ATGGAACTACCAGCTACTGGTGGTCCCTTTAAAATGCCTTCATTGATTTAAACATATAAT GTACATAGGATGTTCCCCACAAGAAAAGTGCAGGCCATGAGAGCTGACTATTACTCACTG TAATGACATTATTATATTTATTTATTACTCAACTTTGAAAGTAATTCAATGATGTCTTCC CTTAAGAACTTTCATGTGTGTAAGTATGACGTAATTGTCTATCAGCTAAATTGACTGTGT TGTTGAGAAATTACCCCATTGAATGTTCTAGGATCTCATTGACCTCTGACTCTGGGTTTT |
STS Forward / Reverse: Foward read
STS:
| CTGGGGTTACTTTCCAGAGCCCTCCNAATTTNNCCCAGGTTTGCTGAATTCCAGTCTGCT CGGGGAAACTCTTTAACCATGCAGCATGGTTCTAGCAGTCAAAAGTGTGGTGTCCTCTTG AGGAGTGATAGCATGAGTGCTTCAAAATGTGTGGGTGTCAACGGAAAATATTGCGACTGA AAAGTGCGTTTTAGGGACAGATAGAAGAGTACCAGACGAGTCATTATCACCCAAAGTCGT AGGCAATTCTTTTACAATGTCTTATGTCAACATAGATTACATAAAAATTTATATTATTTA TTGGATCCAAACAAATTCAGAGTAGTCCATAAAGTTCNG |
STS Forward / Reverse: Reverse read
STS:
| TGGATCCAATAAATAATATAAATTTTTATGTAATCTATGTTGACATAAGACATTGTAAAA GAATTGCCTACGACTTTGGGTGATAATGACTCGTCTGGTACTCTTCTATCTGTCCCTAAA ACGCACTTTTCAGTCGCAATATTTTCCGTTGACACCCACACATTTTGAAGCACTCATGCT ATCACTCCTCAAGAGGACACCACACTTTTGACTGCTAGAACCATGCTGCATGGTTAAAGA GTTTCCCCGAGCAGACTGGAATTCAGCAAACCTG |
STS Forward / Reverse: Reverse read
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-waseComment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
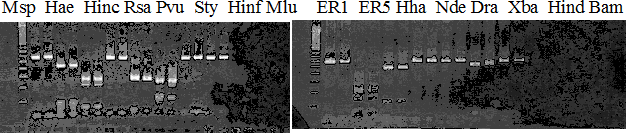 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Rsa1Rank as CAPS marker: 3 or more alleleic model
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Resriction enzyme: Rsa1
Rank as CAPS marker: Others
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Experiments: CAPS pattern of Citrus collection
Experiments: Segregation pattern
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese AcidlessRestriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: NC
Comment: PCR_condition: 56C aneal
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Rsa1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Rsa1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Rsa1
Comment: PCR_condition: 56C aneal
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
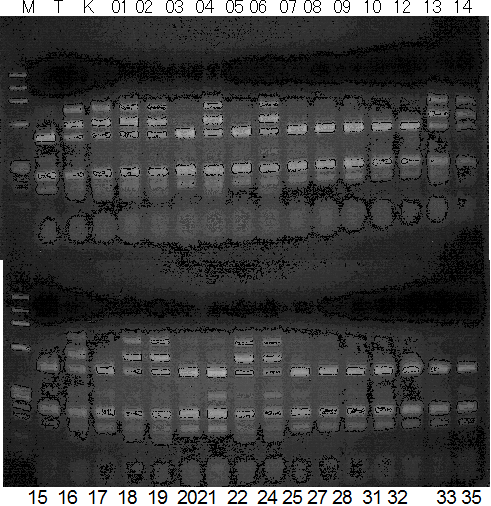 |
Population name: Kiyomi x Okitsu41gou
Restriction enzyme: Rsa1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10018252mScaffold: scaffold_2
Start: 27774863
End: 27778271
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00745:mRNA_6.1Scaffold: C_unshiu_00745
Start: 48150
End: 62370
Gene function: Hypothetical protein
AGI map info (Shimada et al. 2014)
AGI LG: AGI-06AGI cM: 60.007
KmMg map info (Omura et al. 2003)
KmMg LG: KmG-06KmMg cM: 22.141
JtTr map info (Omura et al. 2003)
JtTr LG: JtR-06SJtTr cM: 5.708
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG: NAT-06SKm/O41 cM: 10.082
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI249DNA sequence around SNP1:
| CTTTGAGTATGAGAAAGTAATGGAACTACCAGCTACTGGTGGTCCCTTTAAAATGCCTTC [A/G]TTGATTTAAACATATAATGTACATAGGATGTTCCCCACAAGAAAAGTGCAGGCCA TGAGA |
DNA sequence around SNP2:
| ACATTGTAAAAGAATTGCCTACGACTTTGGGTGATAATGACTCGTCTGGTACTCTTCTAT [T/C]TGTCCCTAAAACGCACTTTTCAGTCGCAATATTTTCCGTTGACACCCACACATTT TGAAG |
DNA sequence around SNP3:
DNA sequence around SNP4: