Locus name: Ov0002
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 58Main product (bp): 1400
Comment on PCR:
F-primer: GAAGAAGACCGGCAAGGTGTT
R-primer: TCTTCCAGGTTGGCGTTGAG
EST info
EST Accession number 1: AU186184EST Accession number 2:
sequence-tagged site (STS seq)
STS:| GATGTCGGGATGAAGAAGAAAGGCGTCGACGAGTTCCCCTTCTGCGTTCACCTCGTCAGT TGGGAGAAGGAGAACGTGTCGAGCGAGGCCTTGGAAGCTGCCCGTATCGCTTGCAACAAG TACATGGCCAAGTACGCCGGCAAGGATGCTTTCCATTTGCGAGTGAGGGTGCACCCCTTC CATGTTCTGCGTATTAACAAGATGCTTTCGTGTGCTGGGGCTGATAGGCTCCAGACTGGA ATGAGGGGTGCTTTTGGAAAGCCCCAAGGAACCTGTGCTCGTGTTGCTATCGGGCAGGTC TTGCTTTCTGTCCGTTGCAAGGATAGTAACAGCCACCATGCTCAGGAGGCCCTTCGCCGT GCTAAGTTCAAGTTTCCTGGTCGTCAAAAGATTATTGTCA |
STS Forward / Reverse: Foward read
STS:
| GGCGTCGACGAGTTCCCCTTCTGCGTTCACCTCGTCAGTTGGGAGAAGGAGAACGTGTCG AGCGAGGCCTTGGAAGCTGCCCGTATCGCTTGCAACAAGTACATGGCCAAGTACGCTGGC AAGGATGCTTTCCATTTGCGAGTGAGGGTGCACCCCTTCCATGTTCTGCGCATTAACAAG ATGCTTTCGTGTGCTGGGGCTGATCGGCTCCAGACTGGAATGAGGGGTGCTTTTGGAAAG CCCCAAGGAACCTGTGCTCGTGTTGCTATCGGGCAGGTCTTGCTTTCTGTCCGTTGCAAG GACAGTAACAGCCACCATGCTCAGGAGGCCCTTCGCCGTGCTAAGTTCAAGTTTCCTGGT CGTCAAAAGATTATTGTCAGCAGGAAATGGTATGCGTTTTTATCCATGACTGCATTCATG TTAAAGTTAACTCGTATTTGCTTGTATTATATGGATTGGCGTGTCACAAGATTAGCAGAT CACATGGATTAAGTTGCCTGGTGCTGGTGTCTCTAGTAGTGAGTTTAAAAAATCTACCCT TATTTAAATTTTTTTCTTTTTTCTTTTTTAAAGAAAAACACAATCAATTAGATGCCTGTT GCTGGTAGTCTCTA |
STS Forward / Reverse: Foward read
STS:
| AGTTCCCCTTCTGCGTTCACCTCGTCAGTTGGGAGAAGGAGAACGTGTCGAGCGAGGCCT TGGAAGCTGCCCGTATCGCTTGCAACAAGTACATGGCCAAGTACGCTGGCAAGGATGCTT TCCATTTGCGAGTGAGGGTGCACCCCTTCCATGTTCTGCGCATTAACAAGATGCTTTCGT GTGCTGGGGCTGATCGGCTCCAGACTGGAATGAGGGGTGCTTTTGGAAAGCCCCAAGGAA CCTGTGCTCGTGTTGCTATCGGGCAGGTCTTGCTTTCTGTCCGTTGCAAGGACAGTAACA GCCACCATGCTCAGGAGGCCCTTCGCCGTGCTAAGTTCAAGTTTCCTGGTCGTCAAAAGA TTATTGTCAGCAGGAAATGGTATGCG |
STS Forward / Reverse: Foward read
STS:
| ACAAGATTAGCAGTTCCCTGGATTAAGTTGCCTGGGCTGGGTCTTTAGTAGTGAGNTTAA AAATCTNCCCTTATTAAATTTTTTCTTTTTTCTTTTTNAAGAAAAAACCCAATCAATTAG ATGCCTGTTGCTGGTAGTCTCTAGGTTTGGGTTTAAAAAATTAACTTCAGGTAGTGTAAT GGAGCTTTAGGGACATATGAGNGTTAGATTGAATTTCTATTTGTTTATTNTAGGCTGATC GNGCTTTTTTTTTTTAAAATACCCATATCTATATGAATGCTCTCTTAAGATCTTGTTTGG GTTTACTTTCTCACTATTGCATAATTATGCTTGTTTTTAGGGGATTTACCAAGTTCAGCC GTGCTGATTATCTGAGGTGGAAGTCGGAGAACAGGATTGTCCCAGATGGTGTGAATGCCA AGGTAACATCAAACTTTATTGGAGTTAAAACATTATTGCATTGCAATTTTGCTTTACTTG AGTTCTTTATAAAATAGTGTTGTCAATCTCTATATTGTGTGTCGAAGCAATCTGCTTTTG CCTCAGTCGATGCATTATTGTTCTGTTCTACTTCTGGAATTTGTTCTCTTCAATTTAGCC TCTCGTGTCCTTTCTTGATTTAGTGTTTATGACTGATGGTATTTGC |
STS Forward / Reverse: Reverse read
STS:
| GAACAAATTCCAGAAGTAGAACAGAACAATAATGCATCGACTGAGGCAAAAGCAGATTGC TTCGACACACAATATAGAGATTGACAACACTATTTTATAAAGAACTCAAGTAAAGCAAAA TTGCAATGCAATAATGTTTTAACTCCAATAAAGTTTGATGTTACCTTGGCATTCACACCA TCTGGGACAATCCTGTTCTCCGACTTCCACCTCAGATAATCAGCACGGCTGAACTTGGTA AATCCCCT |
STS Forward / Reverse: Reverse read
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-waseComment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
 |
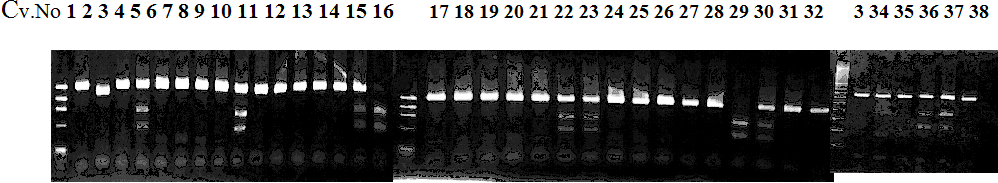
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Hinf1Rank as CAPS marker: Available marker
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
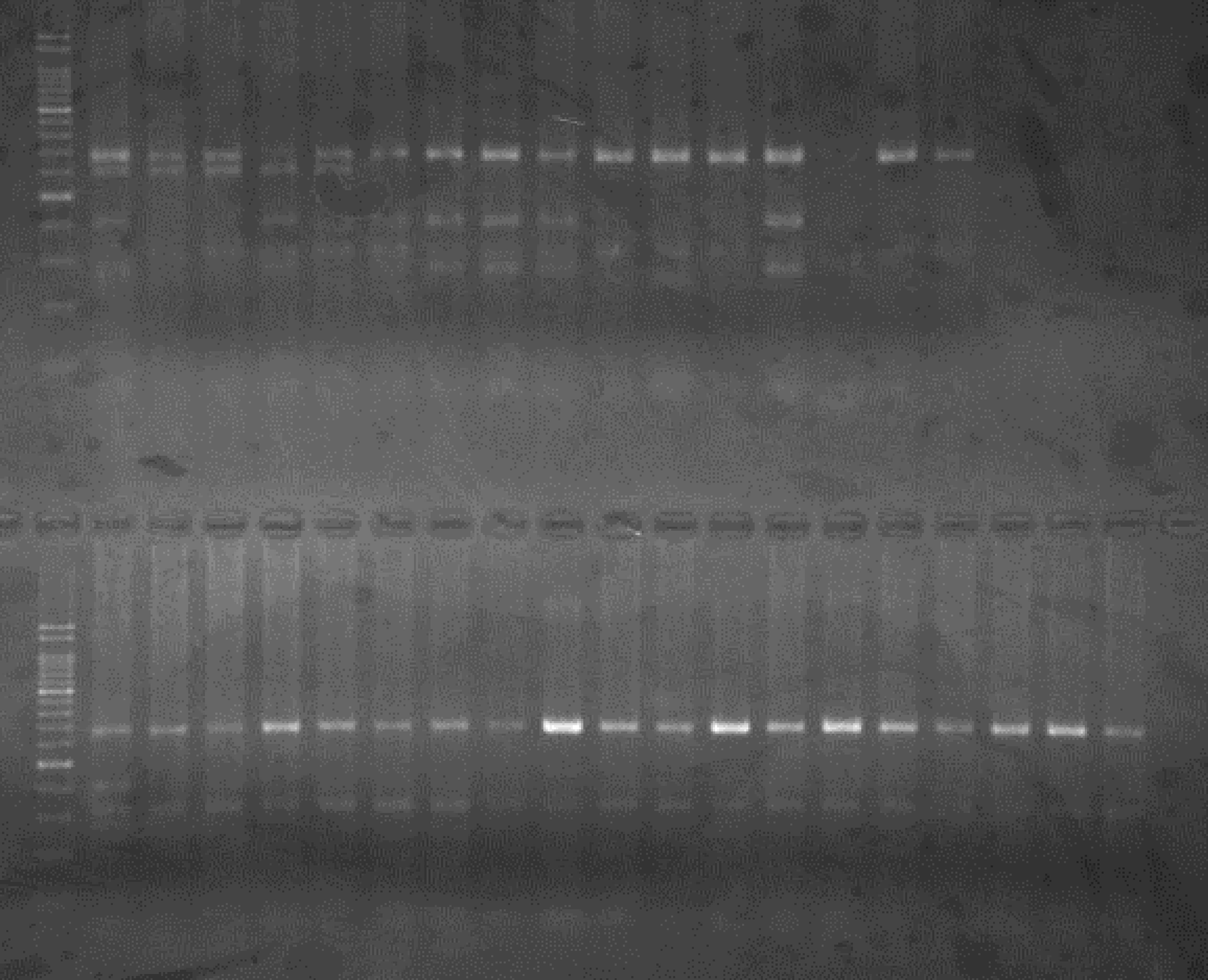
Experiments: CAPS pattern of Citrus collection
Rsetriction enzyme: Hinf1Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
Rsetriction enzyme: Msp1
Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
Experiments: Segregation pattern
Population name: Kiyomi x Okitsu41gouRestriction enzyme: Hinc2
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese Acidless
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10032770mScaffold: scaffold_4
Start: 2422861
End: 2424838
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00979:mRNA_13.2Scaffold: C_unshiu_00979
Start: 50508
End: 52478
Gene function: 60S ribosomal protein L10
AGI map info (Shimada et al. 2014)
AGI LG: AGI-07AGI cM: 13.657
KmMg map info (Omura et al. 2003)
KmMg LG:KmMg cM:
JtTr map info (Omura et al. 2003)
JtTr LG: JtR-07JtTr cM: 68.221
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG: RP-07Rp/Sa cM: 82.035
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI110DNA sequence around SNP1:
| GGCAAGGATGCTTTCCATTTGCGAGTGAGGGTGCACCCCTTCCATGTTCTGCG[T/C]AT TAACAAGATGCTTTCGTGTGCTGGGGCTGAT |
DNA sequence around SNP2:
| AAGTAAAGCAAAATTGCAATGCAATAA[T/C]GTTTTAACTCCAATAAAGTTTGATGTTA CCTTGGCATTCACACCATCTGGGACAATCCTG |
DNA sequence around SNP3:
DNA sequence around SNP4: