Locus name: Ov0020
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 58Main product (bp): 2200
Comment on PCR:
F-primer: AGCTGAGATGGCACTGGT
R-primer: TGGACATAAGATTTGGCAACT
EST info
EST Accession number 1: AU186345EST Accession number 2: DC893762
sequence-tagged site (STS seq)
STS:| TGTGAAACCTAATGTAAGATCTTGAATTGCATGCTAAATGTGTTTTTATAAGTCGCATGA CATAAAGGATACTTTTCTAGGTATCAGTAAGTTCATATATTCTTCTCCTGATAATTTCAT AATTGTTCCTGCAGGTGTATGTCACAATGTATCTCAATGACTGCCAGCGTACTGCTTTCT ATGAGGGCATTGGACTAGACACCAAAGAATTTGACATGCATGTCATCATTGAGGTTAGTC TGTGGCTGAGCTTGTTAGTGTGCTTTTTACAGAAGTTTATGAAGATTTCTGTGCTGCNAA TCAATTGTCAATATGTAGAACTTGACAGCACATTGTGTACCAGCCAAGTGAGCCAACNAA GCTCACTTTAACAATATCCTGCTTTTCTTCATTTGCATTTTNAGTTTTAGTAAGACATCT AACAGTGTCAGTGTCATATCTATATGACCTAGGAAAAGAGGACAGAATTACCATTTTTTA TGATACTTGAACTTGTGTATCTGAAATTCTTTTTCTACAGACCAATCGCACAACGGCTAG AATTTTCCCTGCTGTGCTGGATGTTGAAAACCCAGAGTTCAAGAGGAGGTTGGATCGGAT GGTAGAGATCAACGAGAGGCTTCTTGCTGTAGGGGCGACCGATTNGACA |
STS Forward / Reverse: Reverse read
STS:
| CGGTCGCCCCTACAGCAAGAAGCCTCTCGTTGATCTCTACCATCCGATCCAACCTCCTCT TGAACTCTGGGTTTTCAACATCCAGCACAGCAGGGAAAATTCTAGCCGTTGTGCGATTGG TCTGTAGAAAAAGAATTTCAGATACACAAGTTCAAGTATCATAAAAAATGGTAATTCTGT CCTCTTTTCCTAGGTCATATAGATATGACACTGACACTGTTAGATGTCTTACTAAAACTA AAAATGCAAATGAAGAAAAGCAGGATATTGTTAAAGTGAGCTTTGTTGGCTCACTTGGCT GGTACACAATGTGCTGTCAAGTTCTACATATTGACAATTGATTTGCAGCACAGAAATCTT CATAAACTTCTGTAAAAAGCACACTAACAAGCTCAGCCACAGACTAACCTCAATGATGAC ATGCATGTCAAATTCTTTGGTGTCT |
STS Forward / Reverse: Reverse read
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-waseComment:
Figure legend:
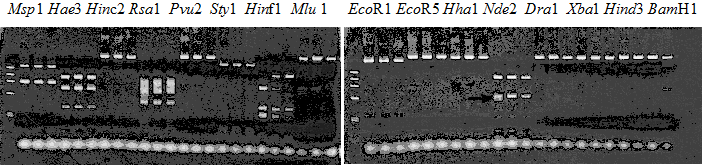 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
 |
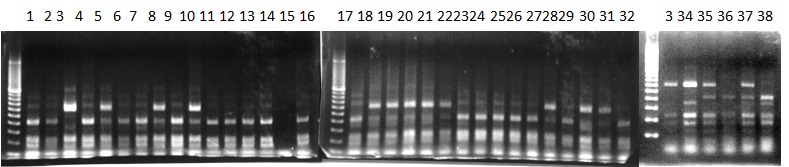
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Hinf1Rank as CAPS marker: Available marker
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
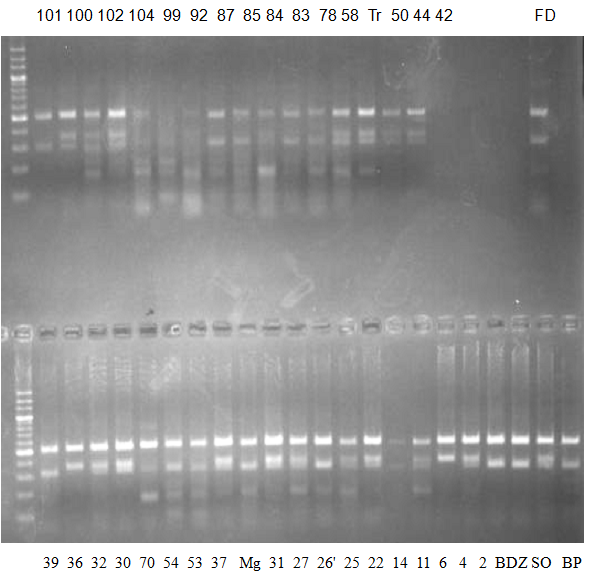
Experiments: CAPS pattern of Citrus collection
Rsetriction enzyme: Hind3Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
Rsetriction enzyme: NC
Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
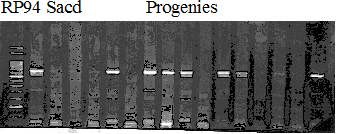
Experiments: Segregation pattern
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese AcidlessRestriction enzyme: Hind3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
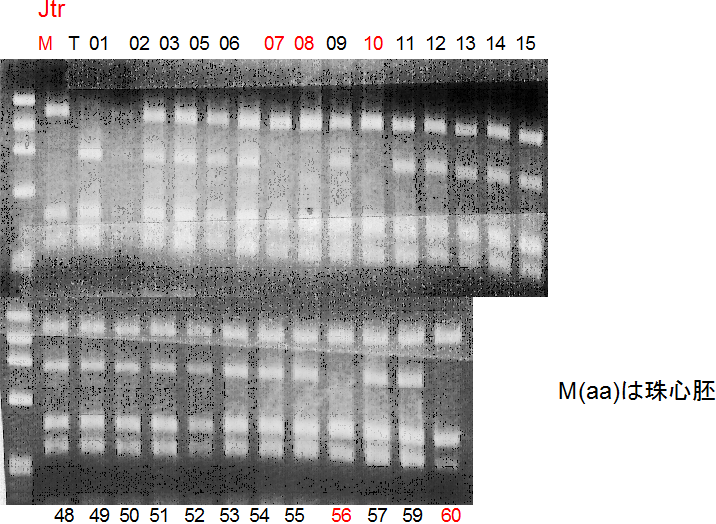 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Okitsu41gou
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10011842mScaffold: scaffold_6
Start: 21598429
End: 21600967
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00037:mRNA_7.2Scaffold: C_unshiu_00037
Start: 37719
End: 40241
Gene function: Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase
AGI map info (Shimada et al. 2014)
AGI LG: AGI-04AGI cM: 55.145
KmMg map info (Omura et al. 2003)
KmMg LG: KmG-04KmMg cM: 22.465
JtTr map info (Omura et al. 2003)
JtTr LG:JtTr cM:
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG: NAT-04Km/O41 cM: 3.806
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG: RP-04Rp/Sa cM: 40.905
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI055DNA sequence around SNP1:
| GTTGGCTCACTTGGCTGGTACACAATGTGCTGTCAAGTTCTACATATTGACAATTGATT[ T/C]GCAGCACAGAAATCTTCATAAACTTCTGTAAAAAGCACACTAACAAGCTCAGCCAC AGAC |
DNA sequence around SNP2:
| ACTGACACTGTTAGATGTCTTACTAAAACT[A/G]AAAATGCAAATGAAGAAAAGCAGGA TATTGTTAAAGTGAGCTT |
DNA sequence around SNP3:
| ATCCAACCTCCTCTTGAACTCTGGGTTTTCAACATCCAGCACAGCAGGGAAAATTCTAGC [T/C]GTTGTGCGATTGGTCTGTAGAAAAAGAATTTCAGATACACAAGTTCAAGTATCAT AAAAA |
DNA sequence around SNP4: