Locus name: Ov0105
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 54Main product (bp): 1200
Comment on PCR:
F-primer: ACATGTGCCGAAAGCCTTAC
R-primer: GGCGGTGATTTATCGTGATTT
EST info
EST Accession number 1: AU186386EST Accession number 2:
sequence-tagged site (STS seq)
STS:| GGATTCCCATGGACATCCGCCATCTTTTTCTGGCTACCATATCAGCAACATCCCATGGAT TTAGCATGGACATCCCCCATCAGATTCATCTTCTCCGACCGAAAGCCGGTTCCCGGAACA ACGATGTTCCCGGCTTGTCCTGCCTGAGTTGGCGACTCGGAGTGGAAACTAATAACGTTA TAGGATGGGCGACTGTTCCCGAAAAATGTGCAGGCTACGTGGGGCATTACATGTTAGGCC AGCAGTACCGCATGGACTCTGAAGTGGTAGCTAATGAGGCCATTCTCTATGCGCAGAGCC TCAAGCTTGCTGGCGACGGCAAGAACATTTGGGTTTTCGACATAGATGAAACCTCGCTAT CTAATTTGCCATATTATGCTAAGAATGGATTCGGGTACGTCCGTGACCGTAAATTAACTT AAACTTCACTTTGTCACCTATATAATTTGGACTTGAGATATATGCCATGATATGTATAGT TTTTAATTTNTTTTTTTT |
STS Forward / Reverse: Foward read
STS:
| CAGCAACATCCCATGGATTTAGCATGGACATCCCCCATCAGATTCATCTTCTCCGACCGA AAGCCGGTTCCCGGAACAACGATGTTCCCGGCTTGTCCTGCCTGAGTTGGCGACTCGGAG TGGAAACTAATAACGTTATAGGATGGGCGACTGTTCCCGAAAAATGTGCAGGCTACGTGG GGCATTACATGTTAGGCCAGCAGTACCGCATGGACTCTGAAGTGGTAGCTAATGAGGCCA TTCTCTATGCGCAGAGCCTCAAGCTTGCTGGCGACGGCAAGAACATTTGGGTTTTCGACA TAGATGAAACCTCGCTATCTAATTTGCCATATTATGCTAAGAATGGATTCGGGTACGTCC GTGACCGTAAATTAACTTAAACTTCACTTTGTCACCTATATAATTTGGACTTGAGATATA TGCCATGATATGTATAGTTTTTAATTT |
STS Forward / Reverse: Foward read
STS:
| TTTTTTTTCAGGGTGAAGCCATACAACCCTACTTTGTTTAATGAATGGGTGAACACCGGA AAAGCACCACCGCTGCCAGAGAGCCTTAAGCTTTACAAAAAATTATTATTGCTTGGAATT AAGATTGTATTCCTCACGGGAAGACCAGAAGATCAGAGAAANGTTACAGAAGCTAATTTA AAGCATGCTGGATTCGACACCTGGGAGAAGCTCATCCTCAAGTAATTAATGATGCGTGTT TTCATCACGTTTATTTTAATGTTTATATGTCAACTAATTAATAACAAGTAAAAATTAATA AATTGGCTGCAGGGGATCATCTCACTCAGGAGAAACAGCAGTGGTGTACAAATCAAGTGA AAGAAAGAAGCTTGAGATGAAAGGGTACAGAATTATTGGAAACATAGGTGATCAATGGAG CGATCTCCTGGGCACCAACGCCGGCAATCGGACGTTCAAGCTGCGGNCCC |
STS Forward / Reverse: Reverse read
STS:
| CGTTGGTGCCCAGGAGATCGCTCCATTGATCACCTATGTTTCCAATAATTCTGTACCCTT TCATCTCAAGCTTCTTTCTTTCACTTGATTTGTACACCACTGCTGTTTCTCCTGAGTGAG ATGATCCCCTGCAGCCAATTTATTAAATTTTTACTTGTTATTAATTAGTTGACATATAAA CATTAAAATAAACGTGATGAAAACACGCATCATTAATTACTTGAGGATGAGCTTCTCCCA GGTGTCGAATCCAGCATGCTTTAAATTAGCTTCTGTAACATTTCTCTGATCTTCTGGTCT TCCCGTGAGGAATACAATCTTAATTCCAAGCAATAATAATTTTTTGTAAAGCTTAAGGCT CTCTGGCAGCGGTGGTGCTTTTCCGGTGTTCACCCATTCATTAAACAAAGTAGGGTTGTA TGGCTTCACCCTGAAAAAAAATTAATT |
STS Forward / Reverse: Reverse read
STS:
| TAATGCCAAGCTCTCGGACTTCGGACTTGCAAAAGATGGTCCTGAGGGTGATAAAACCCA TGTATCAACCCGTGTGATGGGGACTTATGGCTATGCAGCCCCAGAATATGTAATGACAGG TACGTGTAACTAAAGTAAAACTCATTTGAATTCTCTCTTCTCCTTTTTCTTCCCATTTTC AATTTAATTTGAAACCAAAATGTTAATGAGGAAAAGGAAGCCTCGGTCGCAGTCTGTCTG CCTTTCACAGCCTGCATTCATTGCACGCTTTTGGTTTGTAACTTAGTAGAATACAGACTT TTAAGGAACTTTGTGGTTCTATTAGAGCTTATCTTAAATCGGACT |
STS Forward / Reverse: Reverse read
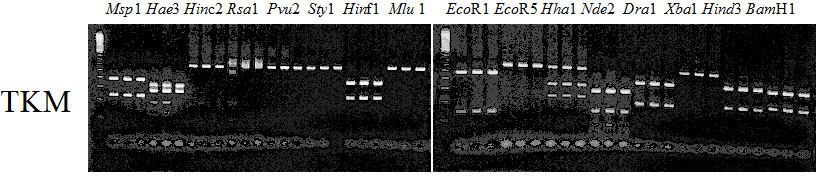
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-waseComment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Hae3Rank as CAPS marker: Disagree by Marco analysis
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
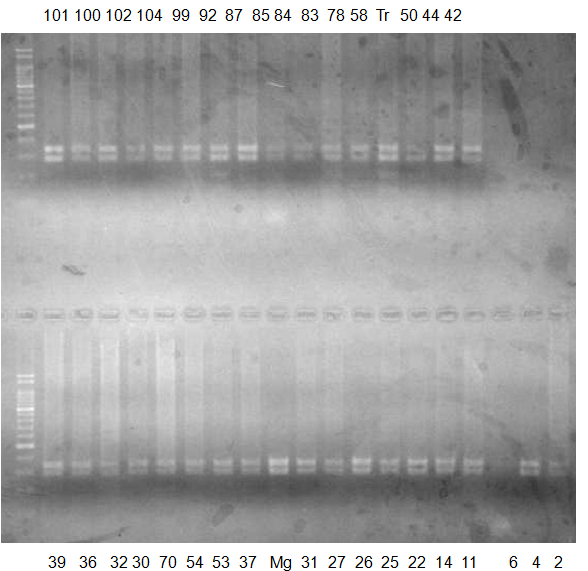
Experiments: CAPS pattern of Citrus collection
Rsetriction enzyme: Hae3Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
Rsetriction enzyme: Hind3
Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
 |
Rsetriction enzyme: Hind3
Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
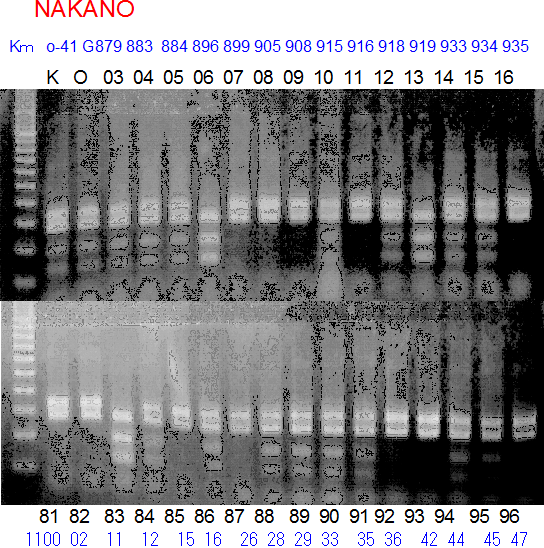
Experiments: Segregation pattern
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese AcidlessRestriction enzyme: Dra1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
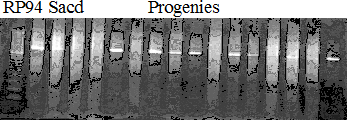 |
Population name: Kiyomi x Okitsu41gou
Restriction enzyme: Hae3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Hae3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Hae3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
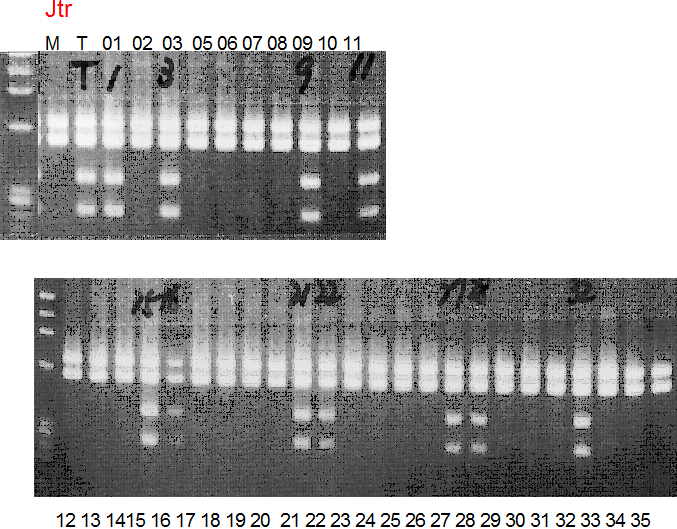 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Hae3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10031362mScaffold: scaffold_4
Start: 23552856
End: 23556415
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00001:mRNA_81.1Scaffold: C_unshiu_00001
Start: 545345
End: 548897
Gene function: Receptor-like kinase plant
AGI map info (Shimada et al. 2014)
AGI LG:AGI cM:
KmMg map info (Omura et al. 2003)
KmMg LG:KmMg cM:
JtTr map info (Omura et al. 2003)
JtTr LG:JtTr cM:
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG: NAT-07Km/O41 cM: 36.023
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG: RP-07Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI126DNA sequence around SNP1:
| ACATTTGGGTTTTCGACATAGATGAAACCTCGCTAT[C/G]TAATTTGCCATATTATGCT AAGAATGGATTCGGGTACGTCCGTGACCGTAAATTAACTTA |
DNA sequence around SNP2:
| CAATAATAATTTTTTGTAAAGCTTAAGGCTCTCTGGCAGCGGTGGTGCTTTTCCGGTGTT [A/C]ACCCATTCATTAAACAAAGTAGGGTTGTATGGCTTCACCCTGAAAAAAAATTAAT T |
DNA sequence around SNP3:
DNA sequence around SNP4: