Locus name: Ov0117
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 60Main product (bp): 1000
Comment on PCR:
F-primer: AGCGAGGGGGATAGCAATACA
R-primer: GACTGACATCCCAAACAGCCA
EST info
EST Accession number 1: AU186413EST Accession number 2: DC893837
sequence-tagged site (STS seq)
STS:| ACTTCCATCAGATAGCATGGGAAGTAATTTCTCAGGAAATCTGCCTGAAAAAACGGCTCA AAATCCGGATAAGGGAATACCAACTGCGAGTGTTGGTTCTCAACATCAAGGCATCTTTCC ACCACTTCATAGCCAGAATGTGCAAATTCCAGCATTTCAACCTCCTTCAGCAATGGGTTA CTACCATCAAAATCCAGTTTCGTGGCCAGCTGCTCCGGCAAATGGATTGATGCCTTTCAC CCATCCCAATCAGTATCTATATACTGGCCCTCTTGGTTATGGATTAAATGGAAACTCTCG CTTGTGCATGCAGTATGGAGGAGCACTTCAGCATGTTGCTACTCCCGTTTTTAATCCTAG CCCGGTGCCGGTTTATCAGTCTATTGCCAAAGCCAACAGCATGGAGAAGCGACCTCACGA TGGTAAGCCAGGTGCACCGCAAGAAGCATTTAACGATACAAATGCAGAGAGGGCTGCTCT GGCCCGGTCACATCTAACTGATGCATTGGCTAAAGGAGAAGGTGGACACCAGAATAATGA TGGCTTTTCCTTGNTTCATTTTGGNGGACCTGTCGGTCTTTCACGGGCTGTAAGTAAATC CTATGCCTTCAAAGATGAG |
STS Forward / Reverse: Foward read
STS:
| TGCATGCACAAGCGAGAGTTTCCATTTAATCCATAACCAAGAGGGCCAGTATATAGATAC TGATTGGGATGGGTGAAAGGCACCAATCCATTTGCCGGAGCAGCTGGCCACGAAACTGGA TTTTGATGGTAGTAACCCATTGCTGAAGGAGGTTGAAATGCTGGAATTTGCACATTCTGG CTATGAAGTGGTGGAAAGATGCTTTGATGTTGAGAACTAACGCTCACAGTTGGTATTCCC TTATCCGGATTTTGAGCCGTTTTTTCAGGCAGATTTCCTGAGAAATTACTTCCCATGCTA TCTGATGGAAGTCCAACAAAAGCTCTTCTTCCCAAAGTCTCTCCTCCATCTGTAATCAGT TTTTTCCCCATTCCAACTTCTTGAAATTCAGAGAAGCCATTTTGAGTGCATGCTGAAGTA TCTCTTCCT |
STS Forward / Reverse: Foward read
STS:
| AAAACTGATTACAGATGNGGAGAGACTTNGGAAGAGAGCTTTTGTTGACTTCCATCAGAT AGCATGGGAAGTAATTTCTCAGGAAATCTGCCTGAAAAAACGGCTCAAAATCCGGATAAG GGAATACCAACTGCGAGTGTTGGTTCTCAACATCAAGGCATCTTTCCACCACTTCATAGC CAGAATGTGCAAATTCCAGCATTTCAACCTCCTTCAGCAATGGGTTACTACCATCAAAAT CCAGTTTCGTGGCCAGCTGCTCCGGCAAATGGATTGATGCCTTTCACCCATCCCAATCAG TATCTATATACTGGCCCTCTTGGTTATGGATTAAATGGAAACTCTCGCTTGTGCATGCAG TATGGAGGAGCACTTCAGCATGTTGCTACTCCCGTTTTTAATCCTAGCCCGGTGCCGGTT TATCAGTCTATTGCCAAAGCCAACAGCATGGAGAAGCGACCTCACGATGGTAAGCCAGGT GCACCGCAAGAAGCATTTAACGATACAAATGCAGAGAGGGCTGCTCTGGCCCGGTCACAT CTAACTGATGCATTGGCTAAAGGAGAAGGTGGACACCAGAATAATGATGGCTTTTCCTTG TTTCATTTTGGTGGACCTGTCGGTCTTTCAACGGGCTGTAAAGTAAATCCTATGCCTTCA AAAGATGAGATTGTTGGGAATTTTTCTTCTCAATTTTCAGCAGATCATGTCGAGAATGAT CATGCTTGCAATAAGAAAGAGACCACAATTGAACAGTACAACTTGTTTGCTGCAAGTAAT GGTAACGGCATACGGTGAC |
STS Forward / Reverse: Reverse read
STS:
| CTTATTGCAAGCATGATCATTCTCGACATGATCTGCTGAAAATTGAGAAGAAAAATTCCC AACAATCTCATCTTTTGAAGGCATAGGATTTACTTTACAGCCCGTTGAAAGACCGACAGG TCCACCAAAATGAAACAAGGAAAAGCCATCATTATTCTGGTGTCCACCTTCTCCTTTAGC CAATGCATCAGTTAGATGTGACCGGGCCAGAGCAGCCCTCTCTGCATTTGTATCGTTAAA TGCTTCTTGCGGTGCACCTGGCTTACCATCGTGAGGTCGCTTCTCCATGCTGTTGGCTTT GGCAATAGACTGATAAACCGGCACCGGGCTAGGATT |
STS Forward / Reverse: Reverse read
STS:
| GATTACAGAT GGAGGAGAGA CTTTGGGAAG ANGAGCTTTT GTTGGACTTC CATCA GATAG CATGGGAAGT AATTTCTCAG GAAATCTGCC TGAAAAAACG GCTCAAAATC CGGATAAGGG AATACCAACT GCGAGTGTTG GTTCTCAACA TCAAGGCATC TTTC CACCAC TTCATAGCCA GAATGTGCAA ATTCCAGCAT TTCAACCTCC TTCAGCAAT G GGTTACTACC ATCAAAATCC AGTTTCGTGG CCAGCTGCTC CGGCAAATGG ATT GATGCCT TTCACCCATC CCAATCAGTA |
STS Forward / Reverse:
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-waseComment:
Figure legend:
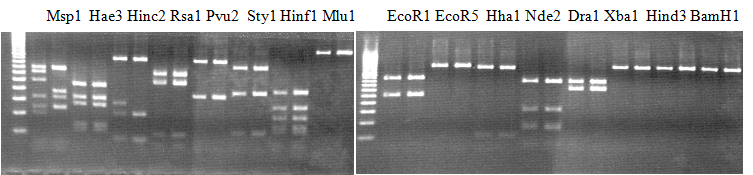 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
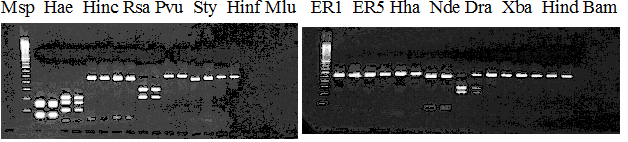 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Dra1Rank as CAPS marker: Available marker
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Experiments: CAPS pattern of Citrus collection
Rsetriction enzyme: Dra1Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
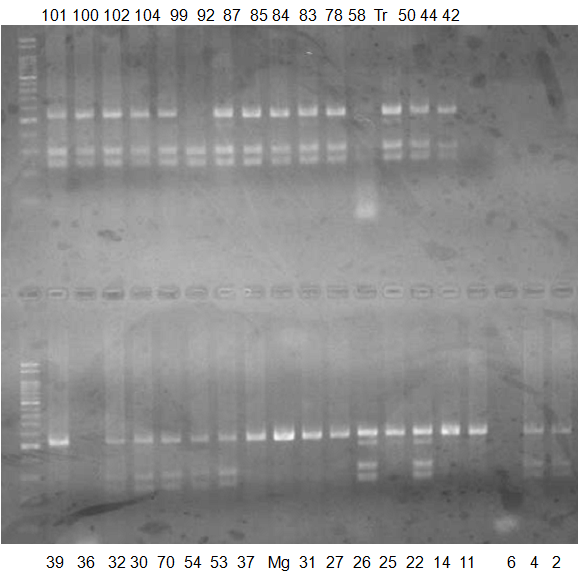 |
Rsetriction enzyme: NC
Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
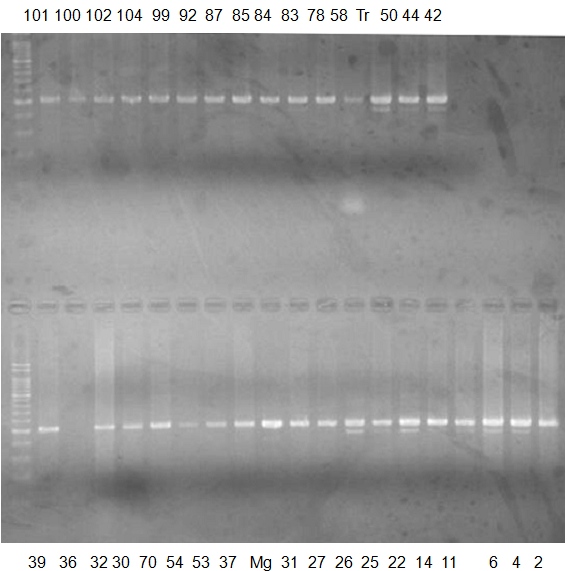 |
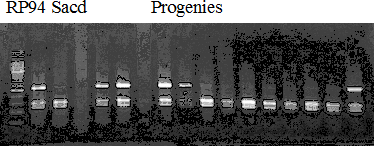
Experiments: Segregation pattern
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese AcidlessRestriction enzyme: Dra1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Dra1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Dra1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10010926mScaffold: scaffold_6
Start: 19105139
End: 19113780
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00089:mRNA_17.1Scaffold: C_unshiu_00089
Start: 153791
End: 161685
Gene function: Hypothetical protein
AGI map info (Shimada et al. 2014)
AGI LG: AGI-04AGI cM: 66.455
KmMg map info (Omura et al. 2003)
KmMg LG:KmMg cM:
JtTr map info (Omura et al. 2003)
JtTr LG:JtTr cM:
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG: RP-04Rp/Sa cM: 28.16
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: Hb, CcCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI040DNA sequence around SNP1:
| GACCGACAGGTCCACCAAAATGAAACAAGGAAAAGCCATCATTATTCTGGTGTCCACCTT [T/C]TCCTTTAGCCAATGCATCAGTTAGATGTGACCGGGCC |
DNA sequence around SNP2:
DNA sequence around SNP3:
DNA sequence around SNP4: