Locus name: Ov0305
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 58Main product (bp): 1250
Comment on PCR:
F-primer: CATCTGGTCCCCACAAGT
R-primer: AGCTTTAGAGAGTGCCAATCA
EST info
EST Accession number 1: AU186464EST Accession number 2: DC893878
sequence-tagged site (STS seq)
STS:| GNATTGCGANCCGTTTGAAATATGCTCTCACATATCGTGAAGTTATTGCTATTCTGATGC AACGACATGTTCTTGTTGATGGGAAGGTTAGGACAGATAAGACCTACCCTGCTGGTTTCA TGGGTATGTGAACTTCTCTCTTTGCAAGTAACTAGATTTTTTTTCCTTTTTTTGGAAATG GTATTCTGTAATCTTGATTTTAGATCTATTAGTTATATAGGAGTAGCAGTTTCTTTTCAT TCTTCTGCCNGGCCTATTCTTTGCAGAATTGTTGGTTTGAGGTACTGAGATTGTTATGTC CTTTTTTTT |
STS Forward / Reverse: Foward read
STS:
| TATGCTCTCACATATCGTGAAGTTATTGCTATTCTGATGCAACGACATGTTCTTGTTGAT GGGAAGGTTAGGACAGATAAGACCTACCCTGCTGGTTTCATGGGTATGTGAACTTCTCTC TTTGCAAGTAACATGATTTTTTTTCCTTTTTTTGGAAATGGTATTCTGTAATCTTGATTT TAGATCTATTAGTTATATAGGAGTAGCAGTTTCTTTTCATTCTTCTGCCTGGCCTATTCT TTGCAGAATTGTTGGTTTGAGGTACTGAGATTGTTATGTCCTTTTTTTT |
STS Forward / Reverse: Foward read
STS:
| CAGAGATGAGGAGGCAAAAGTATGTCTATAACTTATAGTTGATTCTCTTTGTACTCTCTG GGTTTTTAATATTAATTTGCTGTGAAATTGCTTGTTTGAATCAAGAATTATTACTGCTGA TAACTTATCATTATGCATGTATGTAGTTGGTNTCTCCAATTCTCTTACAAACTTGTAGTG CATATTTTCCATTNTTTGCATATTCATCACTAATGAATATCACATGTTAACATCTCGAGT CTCTTTAAATTTATATGAATTTTCTGTAATGCAGTTCAAGCTTTGCAAAGTTAGATCAGT CCAGTTTGGCCAGAAAGGTATCCCATACATCAACACCTATGATGGGCGTACCATCCGCTA CCCTGACCCACTCATCAAAGCCAATGATACCATCAAACTTGACTTAGAAGAGAACAAGAT CACTGACTTCATCAAGTTTGATGTTGGGAATATTGTCATGGTCACTGGGGGTAGAAACAG AGGTCGAGTTGGAATCATCAAGAACAGGGAAAAGCATAAGGGAAGTTTTGAGACCATCCA TATTCAAGATGCCCTTGGTCATGAGTTTGCAACTCGNCTAGGCAATGTGTTCACCATCGG GAAGGGTTCAAAACCATGGGTGTCACTTCCTAAGGGCAAGGGTATTAAGTTGTCCATCAT TGAGGAAGCTAGGAAGAGACAGGCTGCCCAGGCTGCTGG |
STS Forward / Reverse: Reverse read
STS:
| GCAGCCTGGGCAGCCTGTCTCTTCCTAGCTTCCTCAATGATGGACAACTTAATACCCTTG CCCTTAGGAAGTGACACCCATGGTTTTGAACCCTTCCCGATGGTGAACACATTGCCTAGA CGAGTTGCAAACTCATGACCAAGGGCATCTTGAATATGGATGGTCTCAAAACTTCCCTTA TGCTTTTCCCTGTTCTTGATGATTCCAACTCGACCTCTGTTTCTACCCCCAGTGACCATG ACAATATTCCCAACATCAAACTTGATGAAGTCAGTGATCTTGTTCTCTTCTAAGTCAAGT TTGATGGTATCATTGGCTTTGATGAGTGGGTCAGGGTAGCGGATGGTACGCCCATCATAG GTGTTGATGTATGGGATACCTTTCTGGCCAAACTGGACTGATCTAACTTTGCAAAGCTTG |
STS Forward / Reverse: Reverse read
STS:
| ATGGGTATGT GAACTTCTCT CTTTGCAAGT AACTNGATTT TTTTTCCTTT TTTTG GAAAT GGTATTCTGT AATCTTGATT TTAGATCTAT TAGTTATATA GGAGTAGCAG TTTCTTTTCA TTCTTCTGCC CGGCCTATTC TTTGCAGAAT TGTTGGTTTG AGGT ACTGAG ATTGTTATGT CCTTTTTTTT CTCT |
STS Forward / Reverse:
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-waseComment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
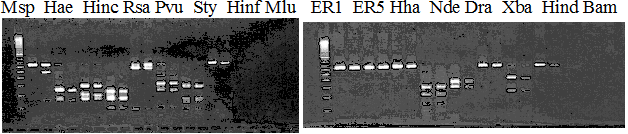 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Msp1Rank as CAPS marker: Disagree by Marco analysis
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
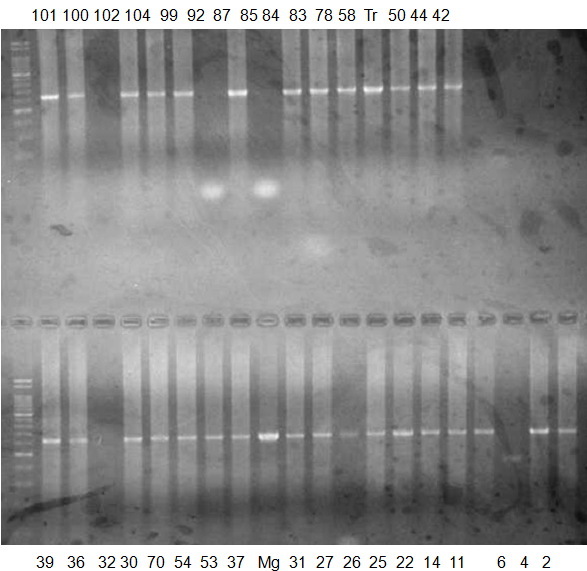
Experiments: CAPS pattern of Citrus collection
Rsetriction enzyme: Msp1Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
Rsetriction enzyme: NC
Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
Rsetriction enzyme: Pvu2
Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
 |
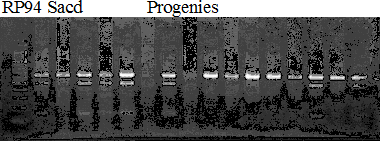
Experiments: Segregation pattern
Population name: Kankitsu Chuukanbohon Nou8gou x Siamese AcidlessRestriction enzyme: Msp1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
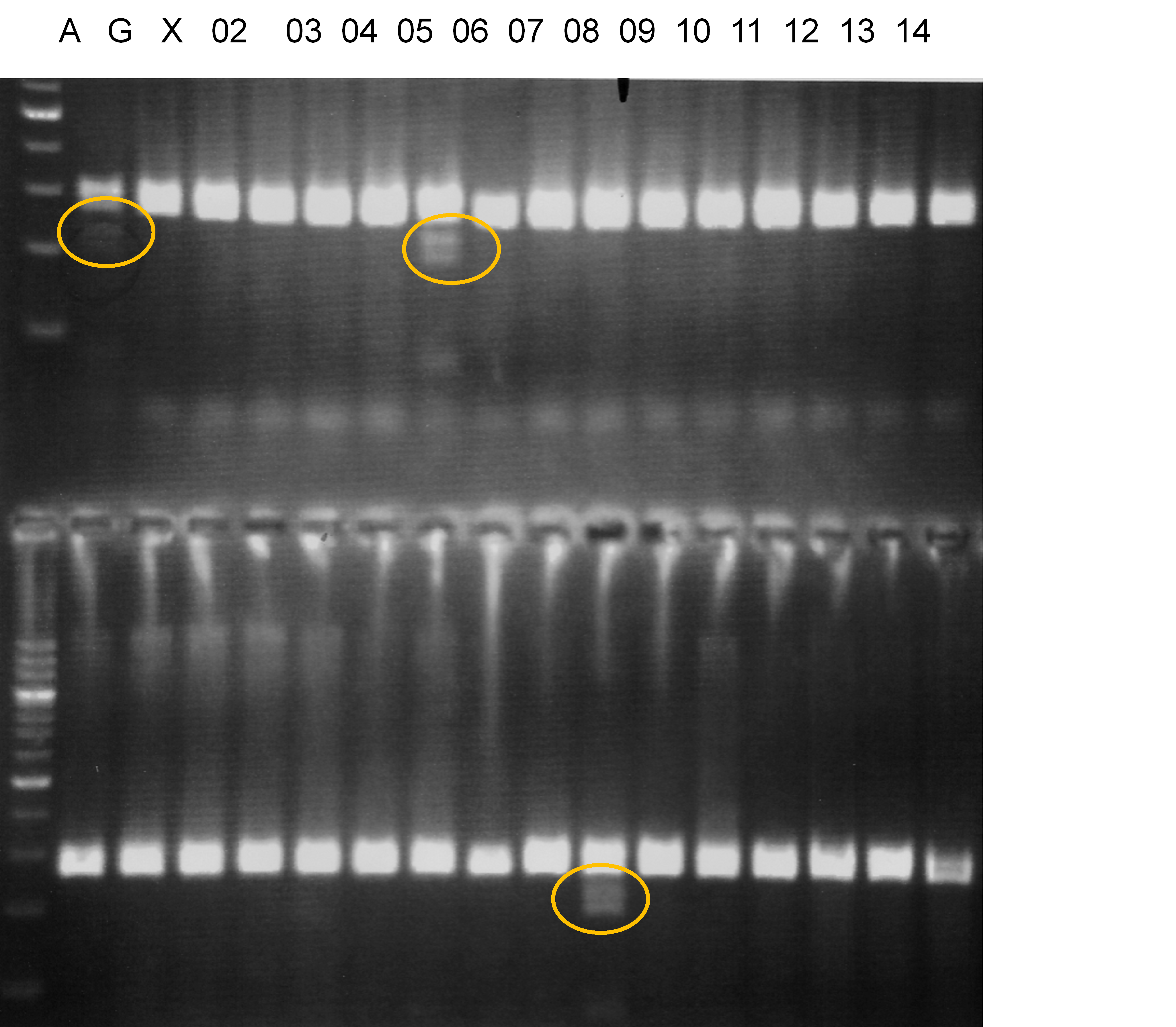
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Msp1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Msp1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
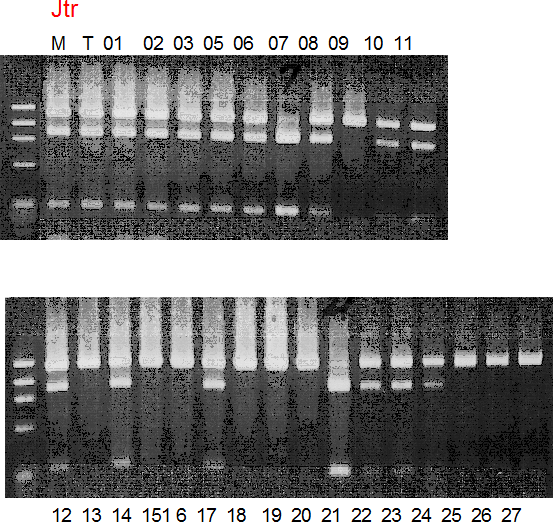 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Msp1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10012499mScaffold: scaffold_6
Start: 16909492
End: 16912088
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00008:mRNA_65.1Scaffold: C_unshiu_00008
Start: 366943
End: 369504
Gene function: 40S ribosomal protein S4
AGI map info (Shimada et al. 2014)
AGI LG: AGI-08AGI cM: 88.433
KmMg map info (Omura et al. 2003)
KmMg LG: KmG-04KmMg cM: 45.755
JtTr map info (Omura et al. 2003)
JtTr LG:JtTr cM:
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG: RP-04Rp/Sa cM: 15.41
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI028DNA sequence around SNP1:
| AATCTTGATTTTAGATCTATTAGTTATATAGGAGTAGCAGTTTCTTTTCATTCTTCTGCC [T/C]GGCCTATTCTTTGCAGAATTGTTGGTTTGAGGTACTGAGATTGTTATGTCCTTTT TTTTC |
DNA sequence around SNP2:
| AATCTTGATTTTAGATCTATTAGTTATATAGGAGTAGCAGTTTCTTTTCATTCTTCTGCC [T/C]GGCCTATTCTTTGCAGAATTGTTGGTTTGAGGTACTGAGATTGTTATGTCCTTTT TTTT |
DNA sequence around SNP3:
DNA sequence around SNP4: