Locus name: Ov0403
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 56Main product (bp): 900
Comment on PCR:
F-primer: CTGTCAAATCCACTCCCCAGA
R-primer: CATAAGCCCAGGCATTGTTG
EST info
EST Accession number 1: DC893934EST Accession number 2: DC893935
sequence-tagged site (STS seq)
STS:| AGCTTGCTTGTGTTGTTGCTTCTTTATCGAGCGTGGCATGCATGTTTGATTTTGGTTACC GATATGCCATTAGTTTTCAGCTTATGTAAAAATAAATAAAGAGATCAGACATAACTTCAA TTTAAAACAAAAAGCTCTAGGCGCAAGAAAANTTGTAATTCTAGCGTAAGCAGTCTCAAG TATCTGATCAAAGAAACCTGTAATCGACAAACAGGTACGGCCCAGACCGCCCAAAGTACT TGGGACCATTCTCTGAGCAAACACCATCATACTTGACTGGTGAGTTCCCTGGTGACTANG GCTGGGATACCGCTGGTTTATCAGCAGACCCCGAGACATTTGCTAAGAACCGTGAGCTAG AGGTGATCCACAGCAGATGGGCAATGCTTGGAGCTCTTGGCTGCGTCTTCCCTGAAATCC TCTCCAAGAATGGAGTGAAGTTCGGCGAAGCAGTTTGGTTCAAGGCTGGCGCTCAAATCT TCTCTGAAGGTGGCCTTGACTATCTTGGCAACCCCAACCTCATTCATGCTCAGAGCATTT TGGCTATCTGGGCCTGCCAGGTTGTGCTCATGGGTTTTGTTGAAGGATACAGAATTGGTG GCGGTCCTCTTGGTGAAGGACTTGACCCACTTTACCCTGGTGGTGCTTTTGACCCACTTG GTTTGGCTGATGACCCCGATCAATTTGCTGAGTTGAAGGTCAAGGAGCTCAAGAATGGCC GTCTGGCTATGTTCTCCATGGTTGGATTCT |
STS Forward / Reverse: Foward read
STS:
| AGCGTGGCATGCATGTTTGATTTTGGTTACCGATATGCCATTAGTTTTCAGCTTATGTAA AAATAAATAAAGAGATCAGACATAACTTCAATTTAAAACAAAAAGCTCTAGGCGCAAGAA AAATTGTAATTCTAGCGTAAGCAGTCTCAAGTATCTGATCAAAGAAACCTGTAATCGACA AACAGGTACGGCCCAGACCGCCCAAAGTACTTGGGACCATTCTCTGAGCAAACACCATCA TACTTGACTGGTGAGTTCCCTGGTGACTAAGGCTGGGATACCGCTGGTTTATCAGCAGAC CCCGAGACATTTGCTAAGAACCGTGAGCTAGAGGTGATCCACAGCAGATGGGCAATGCTT GGAGCTCTTGGCTGCGTCTTCCCTGAAATCCTCTCCAAGAATGGAGTGAAGTTCGGCGAA GCAGTTTGGTTCAAGGCTGGCGCTCAAATCTTCTCTGAAGGTGGCCTTGACTATCTTGGC AACCCCAACCTCATTCA |
STS Forward / Reverse: Foward read
STS:
| AGCGTGNCATGCATGTTTGATTTTGGTTACCGATATGCCATTAGTTTTCAGCTTATGTAA AAATAAATAAAGAGATCAGACATAACTTCAAGTTAAAACAAAAAGCTCTAGGCGCAAGAA AAATTGTAATTCTAGCGTAAGCAGTCTCAAGTATCTGATCAAAGAAACCTGTAATCGACA AACAGGTACGGCCCAGACCGCCCAAAGTACTTGGGACCATTCTCTGAGCAAACACCATCA TACTTGACTGGTGAGTTCCCTGGTGACTANGGCTGGGATACCGCTGGTTTATCAGCAGAC CCCGAGACATTTGCTAAGAACCGTGAGCTAGAGGTGATCCACAGCAGATGGGCAATGCTT GGAGCTCTTGGCTGCGTCTTCCCTGAAATCCTCTCCAAGAATGGAGTGAAGTTCGGCGAA GCAGTTTGGTTCAAGGCTGGCGCTCAAATCTTCTCTGAAGGTGGCCTTGACTATCTTGGC AACCCCAACCTCATTCATGCTCAGAGCATTTTGGCTATCTGGGCCTGCCAGGTTGTGCTC ATGGGTTTTGTTGAAGGATACAGAATTGGTGGCGGTCCTCTTGGTGAAGGACTTGACCCA CTTTACCCTGGTGGTGCTTTTGACCCACTTGGTTTGGCTGATGACCCCGATCAATTTGCT GNTTTGAAGGTCAAGGAGCTCAAGAA |
STS Forward / Reverse: Reverse read
STS:
| CAAAAGCACCACCAGGGTAAAGTGGGTCAAGTCCTTCACCAAGAGGACCGCCACCAATTC TGTATCCTTCAACAAAACCCATGAGCACAACCTGGCAGGCCCAGATAGCCAAAATGCTCT GAGCATGAATGAGGTTGGGGTTGCCAAGATAGTCAAGGCCACCTTCAGAGAAGATTTGAG CGCCAGCCTTGAACCAAACTGCTTCGCCGAACTTCACTCCATTCTTGGAGAGGATTTCAG GGAAGACGCAGCCAAGAGCTCCAAGCATTGCCCATCTGCTGTGGATCACCTCTAGCTCAC GGTTCTTAGCAAATGTCTCGGGGTCTGCTGATAAACCAGCGGTATCCCAGCCATAGTCAC CAGGGAACTCACCAGTCAAGTATGATGGTGTTTGCTCAGAGAATGGTCCCAAGTACTTTG GGCGGTCTGGGCCGTACCTGTTTGTCGATTACAGGTTTCTTTGATCAGATACTTGAGACT |
STS Forward / Reverse: Reverse read
STS:
| TACCGATATG CCATTAGTTT TCAGCTTATG TAAAAATAAA TAAAGAGATC AGACA TAACT TCAANTTAAA ACAAAAAGCT CTAGGCGCAA GAAAAATTGT AATTCTAGCG TAAGCAGTCT CAAGTATCTG ATCAAAGAAA CCTGTAATCG ACAAACAGGT ACGG CCCAGA CCGCCCAAAG TACTTGGGAC CATTCTCTGA GCAAACACCA TCATACTTG A CTGGTGAGTT CCCTGGTGAC TATGGCTGGG ATACCGCTGG TTTATCAGCA GAC CCCGAGA CATTTGCTAA GAACCGTGAG CTAGAGGTGA TCCACAGCAG ATGGGCAA TG CTTGGAGCTC TTGGCTGCGT CTTCCCTGAA |
STS Forward / Reverse:
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-waseComment:
Figure legend:
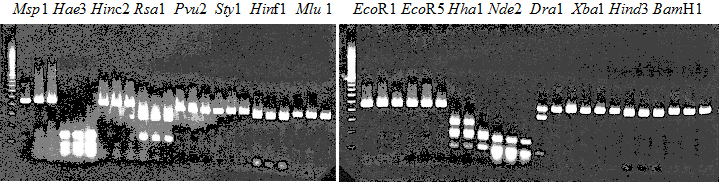 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
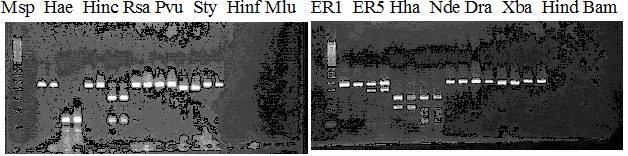 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Hha1Rank as CAPS marker: Available marker
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
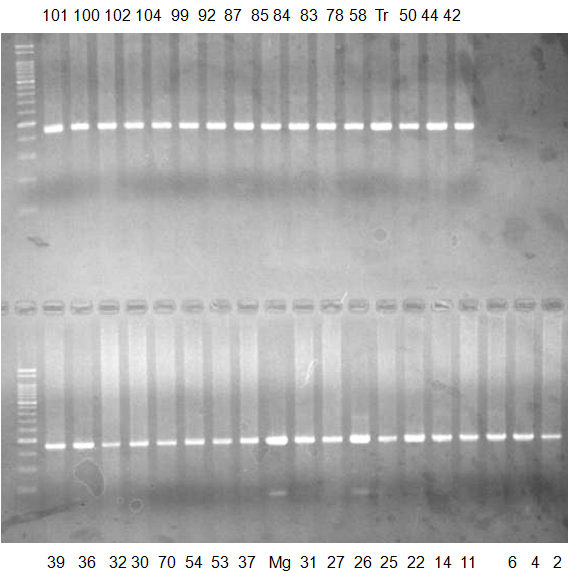
Experiments: CAPS pattern of Citrus collection
Rsetriction enzyme: Hha1Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
Rsetriction enzyme: NC
Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
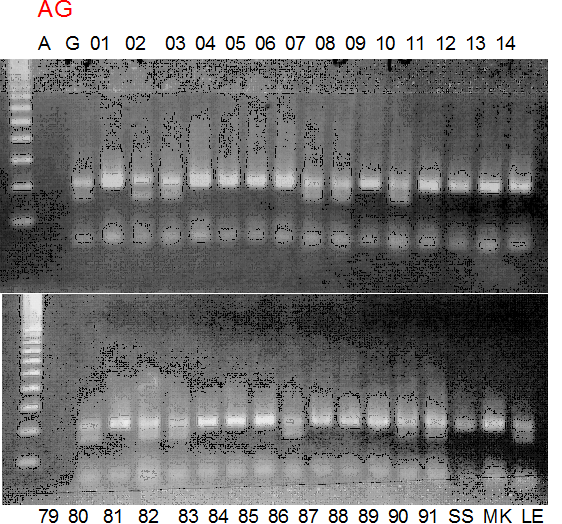
Experiments: Segregation pattern
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)Restriction enzyme: Rsa1
Comment: PCR_condition: 56C aneal
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Rsa1
Comment: PCR_condition: 56C aneal
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Rsa1
Comment: PCR_condition: 56C aneal
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10016280mScaffold: scaffold_2
Start: 23891814
End: 23893095
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00081:mRNA_50.1Scaffold: C_unshiu_00081
Start: 296655
End: 297936
Gene function: Chlorophyll a/b binding protein
AGI map info (Shimada et al. 2014)
AGI LG: AGI-06AGI cM: 61.779
KmMg map info (Omura et al. 2003)
KmMg LG: KmG-06KmMg cM: 12.61
JtTr map info (Omura et al. 2003)
JtTr LG: JtR-06SJtTr cM:
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI136DNA sequence around SNP1:
| CTTGGGACCATTCTCTGAGCAAACACCATCATACTTGACTGGTGAGTTCCCTGGTGACTA [A/T]GGCTGGGATACCGCTGGTTTATCAGCAGACCCCGAGACATTTGCTAAGAACCGTG AGCTA |
DNA sequence around SNP2:
| GATATGCCATTAGTTTTCAGCTTATGTAAAAATAAATAAAGAGATCAGACATAACTTCAA [T/G]TTAAAACAAAAAGCTCTAGGCGCAAGAAAA |
DNA sequence around SNP3:
| CGTCGATCGGGAAGGTCACCCTGTTTGCTACAACATTTATGGGGTGTTTGAGAGTGATGA [A/G]CTTTATCAGAAGACTTTTGGTACTGAGGAGAAGCGTGGCCAGTTCTTGAGATGGA GGCTG |
DNA sequence around SNP4: