Locus name: Ov0513
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 58Main product (bp): 750
Comment on PCR:
F-primer: TGGTTTACATCTGCGAGTG
R-primer: AAATAAAGTGACAAGGCAAAA
EST info
EST Accession number 1: AU186200EST Accession number 2: DC894061
sequence-tagged site (STS seq)
STS:| CGTTTTGTGAACCGCGTAGTTGCCTCTATGTTCAACACGGTTTCAAACAAGAAGATTGCC GTTCTGGGATTTGCCTTCAAGAAGGATACCGGTGACACCAGAGAGACCCCAGCCATTGAT GTCTGCAAGGGGCTTTTGGGAGACAAAGCTCGGTTGAGCATATACGACCCACAGGTCACC GAGGACCAGATCCAGAGGGACCTTACAATGAACAAGTTTGACTGGGACCATCCTCTTCAC CTCCAGCCAATGAGTCCCACCACGGTGAAGCAAGTGAGCGTGGTCTGGGATGCCTATGAA GCAACAAAAGATGCCCATGGTGTCTGCATCTTGACCGAGTGGGATGAGTTCAAGACTCTT GACTATCAGAGGATATATGACAACATGCAGAAACCAGCATTCGTGTTTGATGGCAGGAAT GTTGTTGATGCCAACAAGTTGAGGGAGATTGGTTTCATCGTGTACTCCATT |
STS Forward / Reverse: Combined sequence of both reads
STS:
| CGTCATTAAGATCAATGATTACCAGAAAAGCCGTTTTGTGAACCGCGTAGTTGCCTCTAT GTTCAACACGGTTTCAAACAAGAAGATTGCCGTTCTGGGATTTGCCTTCAAGAAGGATAC CGGTGACACCAGAGAGACCCCAGCCATTGATGTCTGCAAGGGGCTTTTGGGAGACAAAGC TCGGTTGAGCATATACGACCCACAGGTCACCGAGGACCAGATCCAGAGGGACCTTACAAT GAACAAGTTTGACTGGGACCATCCTCTTCACCTCCAGCCAATGAGTCCCACCATGGTGAA GCAAGTGAGCGTGGTCTGGGATGCCTATGAAGCAACAAAAGATGCCCATGGTGTCTGCAT CTTGACCGAGTGGGATGAGTTCAAGACTCTTGACTATCAGAGGATATATGACAACATGCA GAAACCAGCATTCGTGTTTGATGGCAGGAATGTTGTTGATGCCAACAAGTTGAGGGAGAT TGGTTTCATCGTGTACTCCATTGGTAAGCCACTCGATCCATGGTTGAAGGACATGCCGGC TGTTGCATAAAAGAGACGCCAATGGGCATGCTCGTGTCATCGGGCATCAGATTATAGATG ATGAAGAAGTCAGTTCAATTCTTTACTATATTTTGATGA |
STS Forward / Reverse: Foward read
STS:
| CGCGTAGTTGCCTCTATGTTCAACACGGTTTCAAACAAGAAGATTGCCGTTCTGGGATTT GCCTTCAAGAAGGATACCGGTGACACCAGAGAGACCCCAGCCATTGATGTCTGCAAGGGG CTTTTGGGAGACAAAGCTCGGTTGAGCATATACGACCCACAGGTCACCGAGGACCAGATC CAGAGGGACCTTACAATGAACAAGTTTGACTGGGACCATCCTCTTCACCTCCAGCCAATG AGTCCCACCATGGTGAAGCAAGTGAGCGTGGTCTGGGATGCCTATGAAGCAACAAAAGAT GCCCATGGTGTCTGCATCTTGACCGAGTGGGATGAGTTCAAGACTCTTGACTATCAGAGG ATATATGACAACATGCAGAAACCAGCATTCGTGTTTGATGGCAGGAATGTTGTTGATGCC AACAAGTTGAGGGAGATTGGTTTCATCGTGTACTCCATTGGTAAGCCACTCGATCCATGG TTGAAGGACA |
STS Forward / Reverse: Foward read
STS:
| CGGTGACACC AGAGAGACCC CAGCCATTGA TGTCTGCAAG GGGCTTTTGG GAGAC AAAGC TCGGTTGAGC ATATACGACC CACAGGTCAC CGAGGACCAG ATCCAGAGGG ACCTTACAAT GAACAAGTTT GACTGGGACC ATCCTCTTCA CCTCCAGCCA ATGA GTCCCA CCATGGTGAA GCAAGTGAGC GTGGTCTGGG ATGCCTATGA AGCAACAAA A GATGCCCATG GTGTCTGCAT CTTGACCGAG TGGGATGAGT TCAAGACTCT TGA CTATCAG AGGATATATG ACAACATGCA |
STS Forward / Reverse:
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-wase/PoncirusComment:
Figure legend:
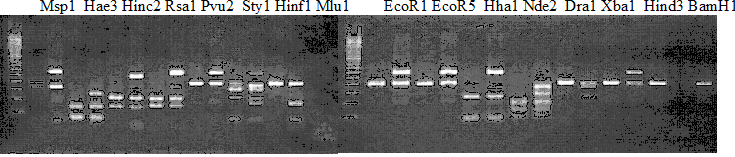 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Msp1Rank as CAPS marker: Others
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
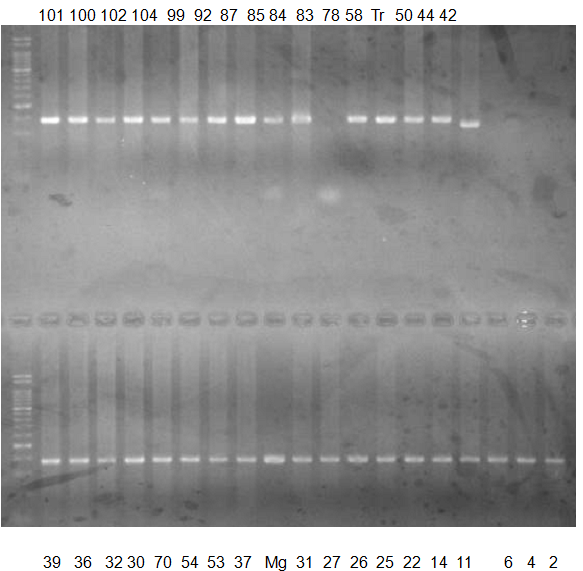
Experiments: CAPS pattern of Citrus collection
Rsetriction enzyme: Msp1Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
Rsetriction enzyme: NC
Comment:
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
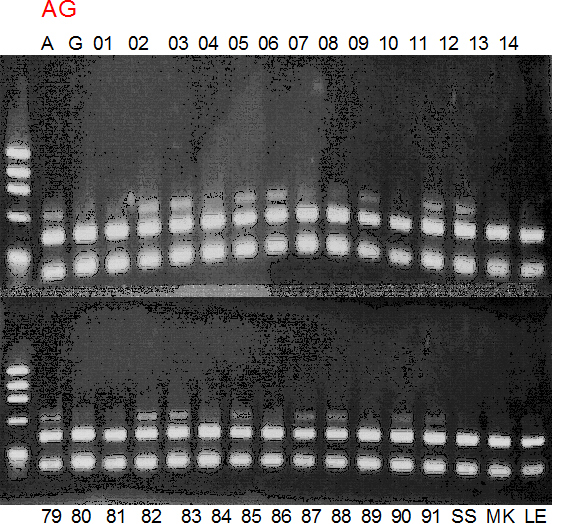
Experiments: Segregation pattern
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)Restriction enzyme: Msp1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
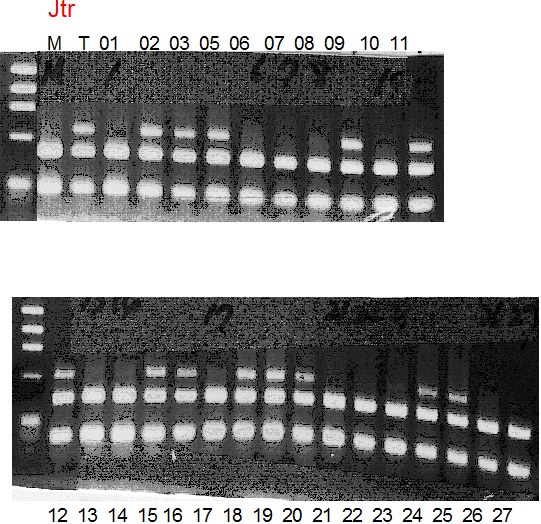
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Msp1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Msp1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
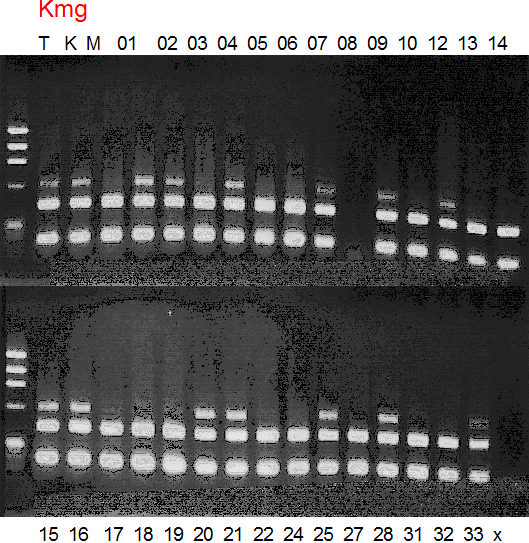 |
Population name: Kiyomi x Okitsu41gou
Restriction enzyme: Msp1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10011622mScaffold: scaffold_6
Start: 25349417
End: 25351949
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00009:mRNA_59.1Scaffold: C_unshiu_00009
Start: 325807
End: 333424
Gene function: Hypothetical protein
AGI map info (Shimada et al. 2014)
AGI LG: AGI-04AGI cM: 38.903
KmMg map info (Omura et al. 2003)
KmMg LG: KmG-04KmMg cM: 5.11
JtTr map info (Omura et al. 2003)
JtTr LG:JtTr cM:
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG: RP-04Rp/Sa cM: 60.491
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping:Cultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1:DNA sequence around SNP1:
DNA sequence around SNP2:
DNA sequence around SNP3:
DNA sequence around SNP4: