Locus name: Tf0022
CAPS PCR info
PCR Anneal-Temp (℃) / Method: TdMain product (bp): 1200
Comment on PCR:
F-primer: CGGCAATGCTACATAACTC
R-primer: AGGGTAGAGAGATGGGCTAAC
EST info
EST Accession number 1: DC891569EST Accession number 2:
sequence-tagged site (STS seq)
STS:| CTATCAGACCGGTATGTTTTATCAGTTATTACCCCAAGTACCAATTCCTTCTACTTAATT TTGTATGACATGTTTTTTAGTTTATTAATTGGATTTTTTCTTCAAAATTTTGTTTATATG GTTTTTTACAGCTTAGATGTTGTTGTGGGCACACCCTTAATATTTCTTTTTTCTGCTGCC CCCCCCCCCCCCTTTGTTTTTACATGCCCACCCTTAATTTAGGCTTTTAGGTCAAGAAAT GCAACATGGTATAAGAGCTAGTTTGACGAAGACATGAAGTTTTTTAGTGGTCACATTCCT TATTGTTAATTAATCGAATTGTGATAATTTGTGTGTATCTATGCTGTTGTCAGGCCCTCA TAATTTGAGCTTTTAGACTAAATAGTGTTTAAAAAGTGTAGAAACTAGATGATCTGTTAT TTTCTAATTACGAAATTGTTTCATCTGACTTTATTTATTTATTTTAAAGAAGAAAAAAAA TTATCTTTACCTCTGGTTAATTCTCTGTTTGTTTTATTTATTTTATATGTTGTATGGGGC TTCCATTGAAAATTTTTATGTTTTGATGCTATCAGGGTGAACTGGATTGTCCTTTCTACT TGAAAACTGGGAGGTGAATATATTGATATTCGTGTTTTCTTTTTTTAATTGAATTATTTA TTTATTTTTTTAAAACGAAAGAATATATAGAGAGAATGAC |
STS Forward / Reverse:
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): A255(Okitsu46gou)/G434(Kankitsu Chuukanbohon Nou5)Comment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Frying dragon/Hyuuganatsu
Comment:
Figure legend:
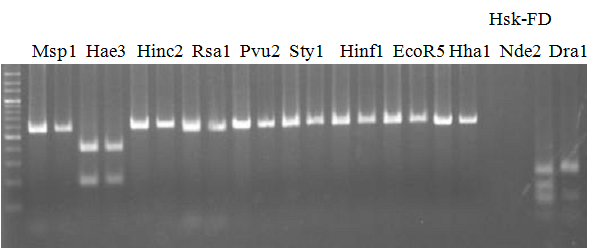 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Hha1Rank as CAPS marker: Disagree by Marco analysis
Comment: DNA sample: New CCN
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
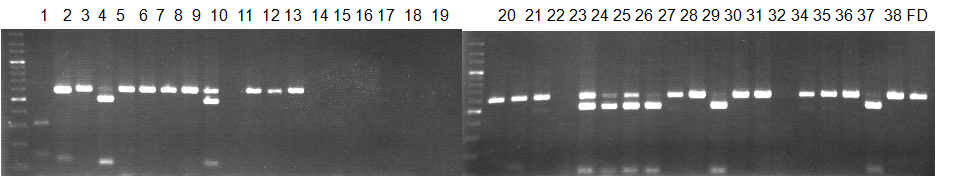 |
Experiments: CAPS pattern of Citrus collection
Rsetriction enzyme: Hha1Comment: DNA sample: New CCN
Figure legend:
CV (No.1 Top) 101 Tahiti, 100 Rangpurlime, 102 Limonia, 104 LisbonL, 99 SweetLime, 92 Hirado, 87 MGF, 85 Kinukawa, 84 Hassaku, 83 Yamamikan, 78 Natsudaidai, 58 Rokugatu, Tr Trovita, 50 WNV, 44 Tankan, 42 Iyo,
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.1 Bottom) 39 HgNt, 36 Ujukitsu, 32 Kawabata, 30 Shunkokan, 70 Yuzu, 54 Henka, 53 Ichang lemon, 37 Kunenbo, Mg Miyagawa, 31 Yatsushiro, 27 Kabuchii, 26 Ponkan, 25 Medet. 22. Dancy, 14 Tachibana, 11 Hirakishu (26' Ohta ponkan)
(No.2 Top) 6 Cleopatra, 4 CQ, 2 Fukuremikan, FD Hiryuu, 124 Largeleaf, 125 Small leaf, 141 CTV-S, 136 Rubidoaux,
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
(No.2 Bottom) FD, Hassaku, Km, F1, RP94, SA, 02, 04, 05, 06, 07, 08, 09, 10, 11, 13
BDZ BenDi Zao, SO Shuto, BP Banpeiyu
 |
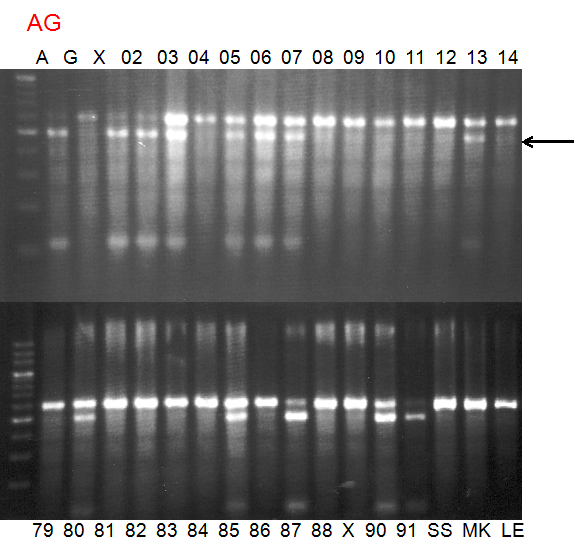
Experiments: Segregation pattern
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)Restriction enzyme: Hha1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
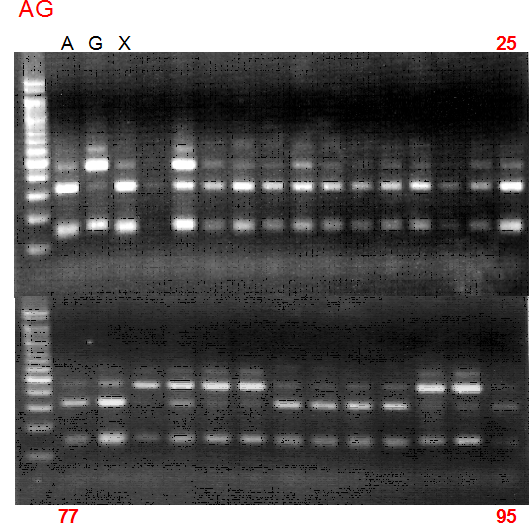
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Hind3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10005681mScaffold: scaffold_9
Start: 31237089
End: 31243866
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00003:mRNA_112.1Scaffold: C_unshiu_00003
Start: 803488
End: 808730
Gene function: Zinc finger CCCH domain-containing protein 32
AGI map info (Shimada et al. 2014)
AGI LG: AGI-03AGI cM: 78.631
KmMg map info (Omura et al. 2003)
KmMg LG:KmMg cM:
JtTr map info (Omura et al. 2003)
JtTr LG:JtTr cM:
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI116DNA sequence around SNP1:
| TGAAGTTTTTTAGTGGTCACATTCCTTATTGTTAATTAATCGAATTGTGATAATTTGTGT [C/G]TATCTATGCTGTTGTCAGGCCCTCATAATTTGAGCTTTTAGACTAAATAGTGTTT AAAAA |
DNA sequence around SNP2:
DNA sequence around SNP3:
DNA sequence around SNP4: