Locus name: Vs0002
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 56Main product (bp): 1200
Comment on PCR:
F-primer: CTAGATGCTGATGTCAAA
R-primer: TAAAGCTTACAACCGACT
EST info
EST Accession number 1: C21910EST Accession number 2:
sequence-tagged site (STS seq)
STS:| GTCTGGACTAGACCCTAGATTGACTTATGNTTTTGCATCCAAGAAAAATTCTAAAGTGAT AACGATGAGACCAACTATTAACACCTGCCTTCATGCTACTAGAAAACATTGTAATACCAG AAATCAGATAACAAAAGGGTGTATAGCTCTATTCATGTGAATTATCATAGATCAGCTGAT CTTTATCTGCCACTACTGGTCCCCTCCTTCCCAGTCACAATTTCTTTATAAAGAGATGTT ACCTTCCAATGAGATAAATTTTGCATAACTGCTCTCCCGCACTTGGTATTAACTGGTTTT TTACACTTGCAGATCCTTGGGCCTGATAGTGGTTGGCAGCCAATTCCACTAACAGATCTT ATAGCAACTGCTGCTGTGAAGAAAGCATACAAGAAGGCCACACTTGTTGTTCATCCCGAC AAGTTACAGCAGCGAGGTGCAAGTATACAGCAGAAGTACACATGTGAAAAGGTTTTTGAT CTTCTAAAGGTGTGTTGTGCAACTTTCTTTCTTTTTTAATTAGCATTTTATAATTTNGTA TAATTAGTTTCTAAAGGAACTTGGGATCTCATGACATTAATTGGTTTCTAATGATATACT AGCAAATTGATTGGGTCTTAGCCTGGTGATGATATCTGCTCTATATATTCAATATAGTGN TATTAAATGAGAAGTATCTTACGTTGAGAGCTCAGTCCCATAATGACGAACTGCATGGGT TAC |
STS Forward / Reverse: Foward read
STS:
| AAAAATTCTAAAGTGATAACGATGAGACCAACTATTAACACCTGCCTTCATGCTACTAGA AAACATTGTAATACCAGAAATCAGATAACAAAAGGGTGTATAGCTCTATTCATGTGAATT ATCATAGATCAGCTGATCTTTATCTGCCACTACTGGTCCCCTCCTTCCCAGTCACAATTT CTTTATAAAGAGATGTTACCTTCCAATGAGATAAATTTTGCATAACTGCTCTCCCGCACT TGGTATTAACTGGTTTTTTACACTTGCAGATCCTTGGGCCTGATAGTGGTTGGCAGCCAA TTCCACTAACAGATCTTATAGCAACTGCTG |
STS Forward / Reverse: Foward read
STS:
| AGCATACAAGAAGGCCACACTTGTTGTTCATCCCGACAAGTTACAGCAGCGAGGTGCAAG TATACAGCAGAAGTACACATGTGAAAAGGTTTTTGATCTTCTAAAGGTGTGTTGTGCAAC TTTCTTTCTTTTTTAAATTAGCATTTTATAAATTTNGTATAATTAGTTTCTAAAGGAACT TGGGATCTCATGACAATTAATTGGTTTCTAAATGATATACTAGCAAATTGATTGGGTCTT AGCCTGGTGATGATAATACTGCTCTATATAATNCAAATAATAGTGTTTATTAAAATGAGA AAGTAATCTTAACGGTTGAGAGTCTCAGTCNCCATTAATNTACGATACTGCAATTGGGTT TTACAACACTCGGTCCGTATCTTATTGGGCCATTACTTTTTCTGTGTTTAGAGGCTCCCT TCTCTTTCATTCTTGATCATGGTCCCCTGAATCTCATTATTTTGTTGGCTGCAGGANGCG TGGAACAGATTCAATGCAGAGGAACGATAAACAGGTTTCCTTCAGCCAAGCATTTTCTTA GGTTAAAGTACGTAATTTATTACCGTATCCTGTAACTAAAAGTAGGAATCGAAGCTTCAC CAACCATTCTTTTCAGACCC |
STS Forward / Reverse: Reverse read
STS:
| GAGAAGGGAGCCTCTAAACACAGAAAAAGTAATGGCCCAATAAGATACGGACCGAGTGTT GTAAAACCCAATTGCAGTATCGTAAATTAATGGAGACTGAGACTCTCAACCGTTAAGATT ACTTTCTCATTTTAATAAACACTATTATTTGNATTATATAGAGCAGTATTATCATCACCA GGCTAAGACCCAATCAATTTGCTAGTATATCATTTAGAAACCAATTAATTGTCATGAGAT CCCAAGTTCCTTTAGAAACTAATTATA |
STS Forward / Reverse: Reverse read
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): A255(Okitsu46gou)/G434(Kankitsu Chuukanbohon Nou5)Comment:
Figure legend:
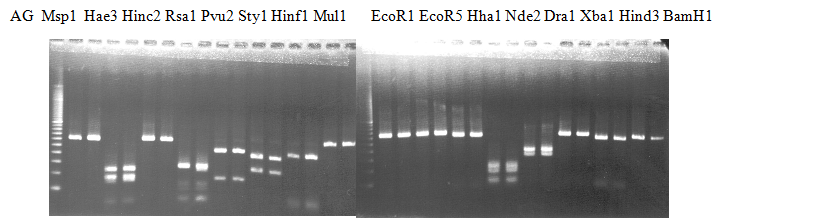 |
Compared cultivars (Left, (Middle), Right): Miyagawa-wase/Trovita orange
Comment:
Figure legend:
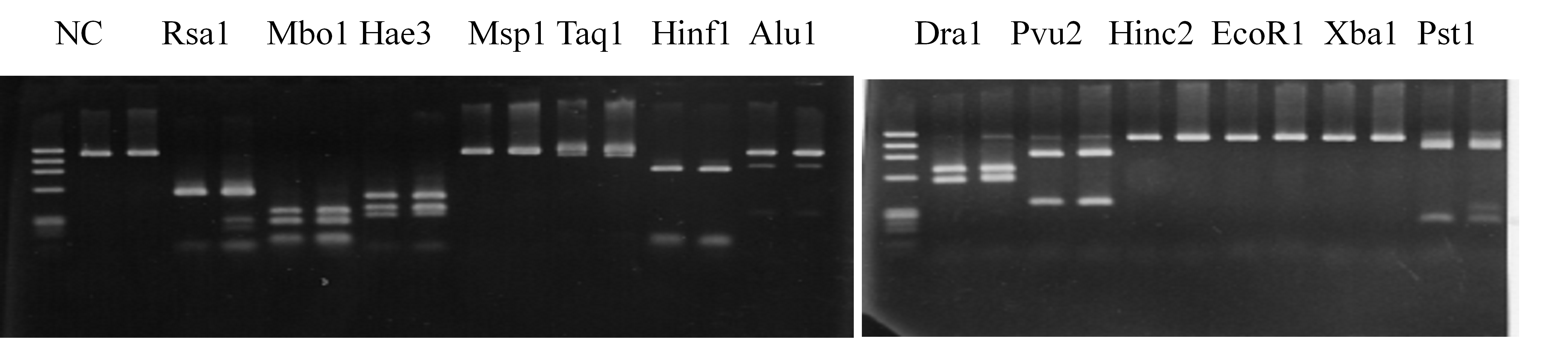 |
Compared cultivars (Left, (Middle), Right): Shiamese acidless/RP-94
Comment:
Figure legend:
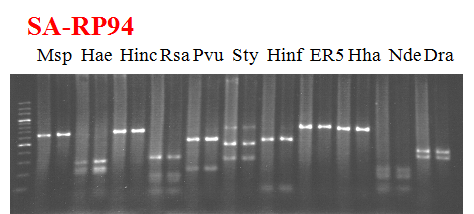 |
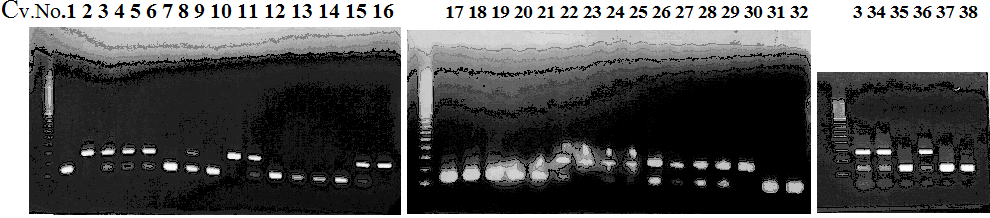
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Hae3Rank as CAPS marker: Others
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Resriction enzyme: Rsa1
Rank as CAPS marker: Disagree by Marco analysis
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Experiments: CAPS pattern of Citrus collection
Experiments: Segregation pattern
Population name: Kiyomi x Miyagawa-waseRestriction enzyme: Hae3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Hae3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Okitsu41gou
Restriction enzyme: Hae3
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
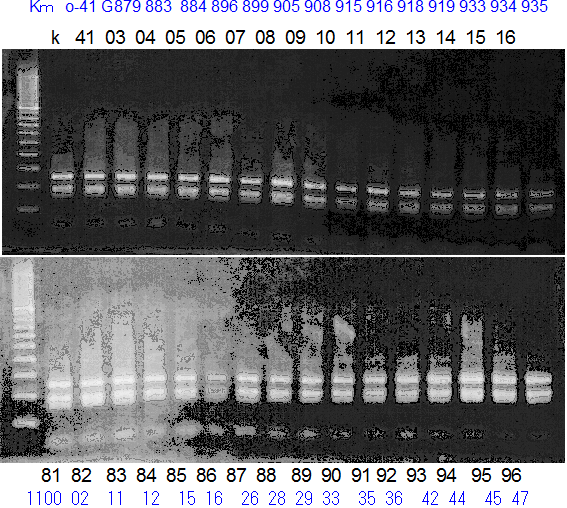 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Rsa1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID: Ciclev10000046mScaffold: scaffold_5
Start: 41184180
End: 41190033
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00127:mRNA_46.1Scaffold: C_unshiu_00127
Start: 341430
End: 344325
Gene function: Hypothetical protein
AGI map info (Shimada et al. 2014)
AGI LG: AGI-09AGI cM: 36.685
KmMg map info (Omura et al. 2003)
KmMg LG: KmG-09KmMg cM: 73.93
JtTr map info (Omura et al. 2003)
JtTr LG: JtR-07JtTr cM: 68.221
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI103DNA sequence around SNP1:
| CTCTATTCATGTGAATTATCATAGATCAGCTGATCTTTATCTGCCACTACTGGTCCCCTC [A/C]TTCCCAGTCACAATTTCTTTATAAAGAGATGTTACCTTCCAATGAGATAAATTTT GCATA |
DNA sequence around SNP2:
| CCTTCCAATGAGATAAATTTTGCATAACTGCTCTCCCGCACTTGGTATTAACTGGTTTTT [A/T]ACACTTGCAGATCCTTGGGCCTGATAGTGGTTGGCAGCCAATTCCACTAACAGAT CTTAT |
DNA sequence around SNP3:
DNA sequence around SNP4: