Locus name: Vs0003
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 54Main product (bp): 950
Comment on PCR:
F-primer: GTACATACTCAAGGCTGG
R-primer: TATTAATACCATCTTCAA
EST info
EST Accession number 1: C21853EST Accession number 2:
sequence-tagged site (STS seq)
STS:| GGTACCGACGGNGGTTCCTGCAAATTTGACAAAAGCAAAATTGCTGCAGCTGTATCTAAT TTCAGTGTTATTTCCTCTGATGAAGATCAAATGGCTGCAAATTTGGTGAAACATGGCCCT CTGGCAGGTAATGTAGCTTCGATACAATTACCTCATATTTCGTTTTCNGTTTCTTGGCTT TTCTCTTCACTGTGAGCTCTCCAAAATAACATTTGGAAAAGTTAGTTAATTAATTAATTT CTTTTGAGATGTTGGTAATTTTTTTATTAAACGGAATGGATAGAATGATGACAGAATTTG TGCTGATCTTGCTGTTTGCTTTTGCAGTGGGTATCAATGCCGTTTGGATGCAAACATATA TTGGAGGAGTTTCATGCCCATACATTTGCGGGAAGTATTTGGATCATGGAGTGCTTATCG TGGGCTATGGATCCTCAGGTTTCGCCCCGATCCGATTCAAGGAGAAGCCTTACTGGATCA TAAAGAACTCCTGGGGAGAGAACTGGGGAGAGAATGGATATTATAAGATCTGCATGGGTC GCAATGTCT |
STS Forward / Reverse: Foward read
STS:
| AAAAGCAAAATTGCTGCAGCTGTATCTAATTTCAGTGTTATTTCCTCTGATGAAGATCAA ATGGCTGCAAATTTGGTGAAACATGGCCCTCTGGCAGGTAATGTAGCTTCGATACAATTA CCTCATATTTCGTTTTCTGTTTCTTGGCTTTTCTCTTCACTGTGAGCTCTCCAAAATAAC ATTTGGAAAAGTTAGTTAATTAATTAATTTCTTTTGAGATGTTGGTAATTTTTTTATTAA ACGGAATGGATAGAATGATGACAGAATTTGTGCTGATCTTGCTGTTTGCTTTTGCAGTGG GTATCAATGCCGTTTGGATGCAAACATATATTGGAGGAGTTTCATGCCCATACATTTGCG GGAAGTATTTGGATCATGGAGTGCTTATCGTGGGCTATGGATCCTCAGGTTTCGCCCCGA TCCGATTCAAGGAGAAGCCTTACTGGATCATAAAGAACTCCTGGGGAGAG |
STS Forward / Reverse: Foward read
STS:
| CGAGAGAAGACTACCCTACACTGGACCGACGGTGGTCCTGCAATTTGACAAAAGCAAAAT TGCTGCAGCTGTATCTAATTTCAGTGTTATTNCNTCTGATGAAGATCAAATGGCTGCAAA TTTGGTGAAACATGGCCCTCTGGCAGGTAATGTAGCTTCGATACAATTACCTCATATTTC GTTTTCNGTTTCTTGGCTTTTCTCTTCACTGTGAGCTCTCCAAAATAACATTTGGAAAAG TTAGTTAATTAATTAATTTCTTTTGAGATGTTGGTAATTTTTTTATTAAACGGAATGGAT AGAATGATGACAGAATTTGTGCTGATCTTGCTGTTTGCTTTTGCAGTGGGTATCAATGCC GTTTGGATGCAAACATATATTGGAGGAGTTTCATGCCCATACATTTGCGGGAAGTATTTG GATCATGGAGTGCTTATCGTGGGCTATGGATCNTCAGGTTTCGCCCCGATCNGNTTCAAG GAGAAGCCTTACTGGATCATAAAGAACTCCTGGGGAGAGAACTGGGGAGAGAATGGATAT TATAAGATCTGCATGGGTCGCAATGTCTGTGGGGTCGACTCCATGGTCTCATCTGTAGCT GCTNTCCATACAACCTCAAGCTAGACATTATGGAGGTTGTGCTAGGCAAGTGGAGCTTNT NTACNNAGATATTATAGGATATCCTTTTAAATATCCGTCTGCAATTATAANGATGCCTAC ATGCNTGGGCTGAGGCATGAACTTTATATGCTCTTGTAATATTTAAGCAT |
STS Forward / Reverse: Reverse read
STS:
| AAGCTCCACTTGCCTAGCACAACCTCCATAATGTCTAGCTTGAGGTTGTATGGATAGCAG CTACAGATGAGACCATGGAGTCGACCCCACAGACATTGCGACCCATGCAGATCTTATAAT ATCCATTCTCTCCCCAGTTCTCTCCCCAGGAGTTCTTTATGATCCAGTAAGGCTTCTCCT TGAA |
STS Forward / Reverse: Reverse read
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): A255(Okitsu46gou)/G434(Kankitsu Chuukanbohon Nou5)Comment:
Figure legend:
 |
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-wase
Comment:
Figure legend:
 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: Hinf1Rank as CAPS marker: Others
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Resriction enzyme: BamH1
Rank as CAPS marker: Available marker
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Resriction enzyme: Hinf1
Rank as CAPS marker: Available marker
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Experiments: CAPS pattern of Citrus collection
Experiments: Segregation pattern
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)Restriction enzyme: BamH1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Miyagawa-wase
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Jutaro unshu x Trovita orange
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
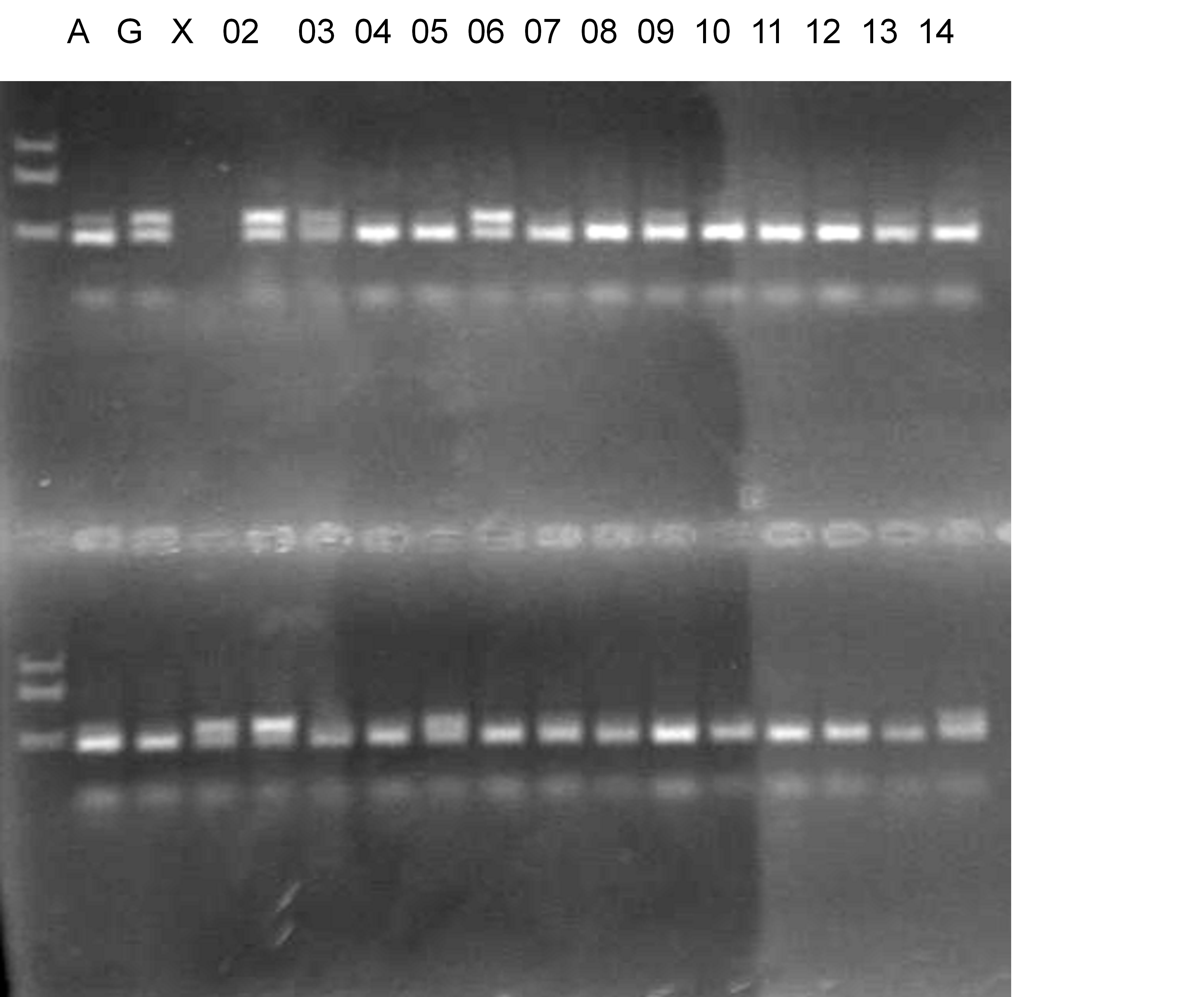 |
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Okitsu41gou
Restriction enzyme: Hinf1
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID:Scaffold:
Start:
End:
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00009:mRNA_117.1Scaffold: C_unshiu_00009
Start: 675718
End: 677501
Gene function: Cysteine proteinase RD19a
AGI map info (Shimada et al. 2014)
AGI LG: AGI-09AGI cM: 35.312
KmMg map info (Omura et al. 2003)
KmMg LG: KmG-09KmMg cM: 72.447
JtTr map info (Omura et al. 2003)
JtTr LG:JtTr cM:
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG: NAT-09Km/O41 cM: 93.467
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1: SI077DNA sequence around SNP1:
| AAGCTCCACTTGCCTAGCACAACCTCCATAATGTCTAGCTTGAGGTTGTATGGA[T/C]A GCAGCTACAGATGAGACCATGGAGTCGACCCCACAGACATTGCGACCCATGCAGATCTT |
DNA sequence around SNP2:
DNA sequence around SNP3:
DNA sequence around SNP4: