Locus name: Vs0014
CAPS PCR info
PCR Anneal-Temp (℃) / Method: 54Main product (bp): 900
Comment on PCR:
F-primer: TATCACCAAAGGAAAACG
R-primer: CAATATAGATAAAAGTCT
EST info
EST Accession number 1: C21868EST Accession number 2:
sequence-tagged site (STS seq)
STS:| TAATTAACTAGCCATGGATCTGCAACAGACCCAGTTAAGGATTCGGAGAAGCAGCCATGG CTGGTTTTTATTTTACAAACAAAAGCTCATAAAAAGACATCCAACAAAGCCAACCACAAA CGATAAATAGTTCTTTTTATTGATACTATAATTTAAACGACCTCACAGCGCTGCTTTATT CTGGGGCATGTTCACGTCTCGTAGTCTTCATTAACAGTTACAAGGGTCGCACTTGCAGTT TGGTCCACACTTGCAGCCGCAGCCGCCCTCGGCTCCGAATCCCATCTCCGACCCCTCAGA GTGCCTGATCCCAAGGAGGGAAAAAAATTTTAAAAAAAATCAAATACAGTCCAAACCTAT CCATCATTTGGGTCCCTGAATTTATATAAAAACAAAAATAATGACACCGTTTGGATTCCT TTTCCATGCAGGCTAATGTAACGTCAGATTGCTTTAAATCTGTTAAATAAAAATCAATAA TTTAAAACACAGTTCTGTAACGACTGCACATTTACGTACA |
STS Forward / Reverse: Combined sequence of both reads
STS:
| GTCCNGAGACACTTAATGATTAATTAACTAGCCATGGATCTGCAACAGACCCAGTTAAGG ATTCGGAGAAGCAGCCATGGCTGGTTTTTATTTTACAAACAAAAGCTCATAAAAAGACAT CCAACAAAGCCAACCACAAACGATAAATAGTTCTTTTTATTGATACTATAATTTAAACGA CCTCACAGCGCTGCTTTATTCTGGGGCATGTTCACGTCTCGTAGTCTTCATTAACAGTTA CAAGGGTCGCACTTGCAGTTTGGTCCACACTTGCAGCCGCAGCCGCCCTCGGCTCCGAAT CCCATCTCCGACCCCTCAGAGTGCCTGATCCCAAGGAGGGAAAAAAATTTTAAAAAAAAT CAAATACAGTCCAAACCTATCCATCATTTGGGTCCCTGAATTTATATAAAAACAAAAATA ATGACACCGTTTGGATTCCTTTTCCATGCAGGCTAATGTAACGTCAGATTGCTTTAAATC TGTTAAATAAAAATCAATAATTTAAAACACAGTTCTGTAACGACTGCACATTTACGTACA TCTTGACAGGAGCAACACCGAGGACGAGGGTCTCGGTGGCTACGGTGGTGCTCTCAAAGG ATCTCAAGTCAGGGTACATGGAGCTACTGCATGTCCAAAAACGAATTAA |
STS Forward / Reverse: Foward read
Experiments: Restriction fragment profile
Compared cultivars (Left, (Middle), Right): Trovita orange/Kiyomi/Miyagawa-waseComment:
Figure legend:
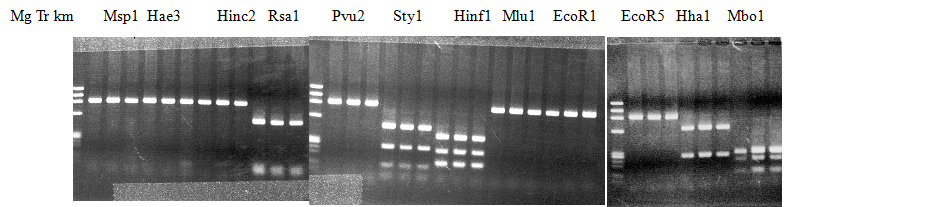 |
Experiments: CAPS pattern of hybrid cultivar
Resriction enzyme: NCRank as CAPS marker: no homozygous cultivar with minor allele
Comment:
Figure legend:
CVName 1. Nagami, 2. Budha's, 3. Miyagawa, 4. Trovita, 5. Kiyomi, 6. Tachibana, 7. Koji, 8. Kinokuni (8' Hirakishu), 9. Mato pumello, 10. Kunip,
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
11. King, 12. Ponkan (12' Ohta ponkan), 13. Dancy, 14. Willowleaf, 15. Clementine, 16. C-haploid, 17. Encore, 18. Amaka, 19. Youkou, 20. Shiranuhi,
21. Harumi, 22. Nishinokaori, 23. Duncan GF, 24. Mineola, 25. Seminol, 26. Seihou, 27. Akemi, 28. Tsunokaori, 29. Ariake, 30. Nankou,
31. Mihocore, 32. Hareyaka, 34. Harehime, 35. Hayaka, 36. Southern Red, 37. Amakusa, 38. Setoka, *. Flying Dragon, SY(Southern Yellow), 40(MK, Mukakukishu)
 |
Experiments: CAPS pattern of Citrus collection
Experiments: Segregation pattern
Population name: A255 (Okitsu46gou) x G434 (Kankitsu Chuukanbohon Nou5gou)Restriction enzyme: NC
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Population name: Kiyomi x Okitsu41gou
Restriction enzyme: NC
Comment:
Figure legend:
A: A255 (Okitu46gou), G: G434 (Kankitsu Chuukanbohon Nou5gou),
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
K or Km: Kiyomi, M or Mg: Miyagawa-wase, MK: Mukaku kisyu,
LE: Lee, O: Okitsu41gou, RP or RP94: Kankitsu Chukanbohon Nou6gou,
Sacd or SA: Siamese Acidless, SW: Sweet spring, T or Tr: Trovita orange,
X: meaningless
 |
Target on C. clementina (v1.0)
Transcript ID:Scaffold:
Start:
End:
Target on C. unshiu (v1.0)
Transcript ID: C_unshiu_00375:mRNA_10.1Scaffold: C_unshiu_00375
Start: 50515
End: 51680
Gene function: Metallothionein
AGI map info (Shimada et al. 2014)
AGI LG:AGI cM:
KmMg map info (Omura et al. 2003)
KmMg LG:KmMg cM:
JtTr map info (Omura et al. 2003)
JtTr LG:JtTr cM:
Km/O41 map info (Nakano et al. 2003)
Km/O41 LG:Km/O41 cM:
Rp/Sa map info (Ohta et al. 2011)
Rp/Sa LG:Rp/Sa cM:
CAPS genotyping (Hybrid cv/Citrus collection)
CAPS genotyping: HbCultivar identification (M: Cultivar identification manual / T: Ninomiya et al. 2015/ K: Nonaka et al. 2017)
Cultivar identification M/T/K:SNP typing by Illumina GoldenGate assay (Fujii et al. 2013)
SNP1:DNA sequence around SNP1:
DNA sequence around SNP2:
DNA sequence around SNP3:
DNA sequence around SNP4: